The Microbiome of Suaeda monoica and Dipterygium glaucum From Southern Corniche (Saudi Arabia) Reveals Different Recruitment Patterns of Bacteria and Archaea
- 1College of Science, Department of Biology, University of Jeddah, Jeddah, Saudi Arabia
- 2Faculty of Fisheries and Food Science, University Malaysia Terengganu, Kuala Nerus, Malaysia
- 3Faculty of Science, Department of Biology, King Abdulaziz University, Jeddah, Saudi Arabia
- 4Department of Biology, College of Sciences, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia
- 5College of Sciences and Arts, Department of Biology, University of Jeddah, Jeddah, Saudi Arabia
- 6Biological Sciences Department, College of Science & Arts, King Abdulaziz University, Rabigh, Saudi Arabia
- 7Faculty of Science, Department of Biochemistry, King Abdulaziz University, Jeddah, Saudi Arabia
- 8Department of Biochemistry, College of Science, Jeddah, Saudi Arabia
- 9Department of Biological Science, King Abdulaziz University, Jeddah, Saudi Arabia
- 10Institute of Food Security and Sustainable Agriculture, Universiti Malaysia Kelantan, Jeli, Malaysia
- 11Department of Agriculture Science, Faculty of Agro-Based Industry, Universiti Malaysia Kelantan, Jeli, Malaysia
Soil and plant interact differently in response to the same stress (e.g., salinity) and recruit certain bacteria. The southern corniche (Saudi Arabia) has limited plant growth, which could be due to the high temperature and salinity. The study aimed to determine the soil microbiome of selected plants and the interactions between soil and these plants. Suaeda monoica and Dipterygium glaucum soil samples were collected from the crust (surface) and rhizosphere, while soil with no plant growth from the nearby area was used as control. High-throughput hypervariable V3–V4 region of the 16S rRNA gene was used to evaluate the shifts in soil microbiome due to growth of plant growth. The analysis detected up to 16% archaeal strains in S. monoica-associated samples, while D. glaucum and control samples contained 100% bacterial strains. The top 10 phyla composition of the soil samples were Proteobacteria, Actinobacteria, Firmicutes, Gemmatimonadota, Bacteroidota, Halobacterota, Cyanobacteria, Cloroflexi, Planctomycetota, and Myxococcota. The V3–V4 region analysis successfully clustered the 5 samples into 3 clusters (control, D. glaucum, and S. monoica) at higher-order classification but not at the species level due to unidentified bacteria. The main differences between soil samples were due to halophyte S. monoica samples containing high amounts of halophilic archaea and halophilic bacteria. This showed that selected plants interacted differently with the soil. EC- and KO-based analyses of functional genes and pathways showed that 5 pathways were specific to control, 11 pathways were observed only in D. glaucum samples, 12 pathways were expressed in S. monoica samples only, and 9 pathways were common in all samples. The study also detected numerous relatively novel genera in high abundance such as Aliifodinibius, Pontibacter, and Lacunisphaera. This showed that the soil in the sampling area is not well explored and that novel species could be isolated from the soil samples and used for future research.
1 Introduction
The southern corniche (Jeddah, Saudi Arabia) is located in the coastal area of the Red Sea and hosts various attractions and tourism spots. In several sections of this long corniche, there is very limited plant growth, which could be due to the high temperature and salinity. The selected sampling area was a good site to explore the soil microbiome and try to understand the interactions between soil and plant. Two plants, namely, Suaeda monoica and Dipterygium glaucum, were found to grow nearby and surrounded by a large empty area.
S. monoica is a well-studied coastal halophyte that grows in marine environments (Devadatha et al., 2018). It was reported to possess various actives such as antiviral, antioxidant, wound healing (Rajathi et al., 2014), phytoremediation (Joshi et al., 2020), and antimicrobial activities (Muthazhagan et al., 2014). As a halophyte, it has also been used in the reclamation of salt-affected agricultural lands due to its ability to absorb sodium chloride (Ayyappan et al., 2013). A previous study on S. monoica in Saudi Arabia reported that this plant is used to treat various other diseases such as rheumatism, paralysis, asthma, and snakebites (Al-Said et al., 2017). A recent study identified novel marine fungi associated with S. monoica in India, which indicates that its microbiome is still under discovery (Devadatha et al., 2018).
D. glaucum Decne. is a monotypic genus with one species belonging to the family Capparidaceae, a slender, shrubby plant with small yellow flowers (Batanouny and Baeshin, 1982; Altwaty et al., 2016; Alzahrani et al., 2020). It is commonly distributed along the Arabian Gulf coast, and Saudi Arabia (in Wadi beds), especially after seasonal rain (Batanouny and Baeshin, 1983). This species suffers from the rarity of water and the very high temperature, which affect its phenotypic characteristics. However, this traditional plant, with multiple medicinal uses, is popular for the treatment of miss-breathing troubles. Previous phytochemical studies on D. glaucum revealed its antioxidant, antimicrobial, and antispasmodic activities (Altwaty et al., 2016; Shaheen et al., 2017).
Hence, this study aimed to compare the bacterial communities in saline soil with limited plant growth and evaluate the shifts in soil microbiome due to the growth of plants such as S. monoica and D. glaucum. The study also evaluated soil samples taken from the crust (surface) as well as rhizosphere of selected plants and compared them with control (no plant growth) samples.
2 Material and Methods
2.1 Site Description
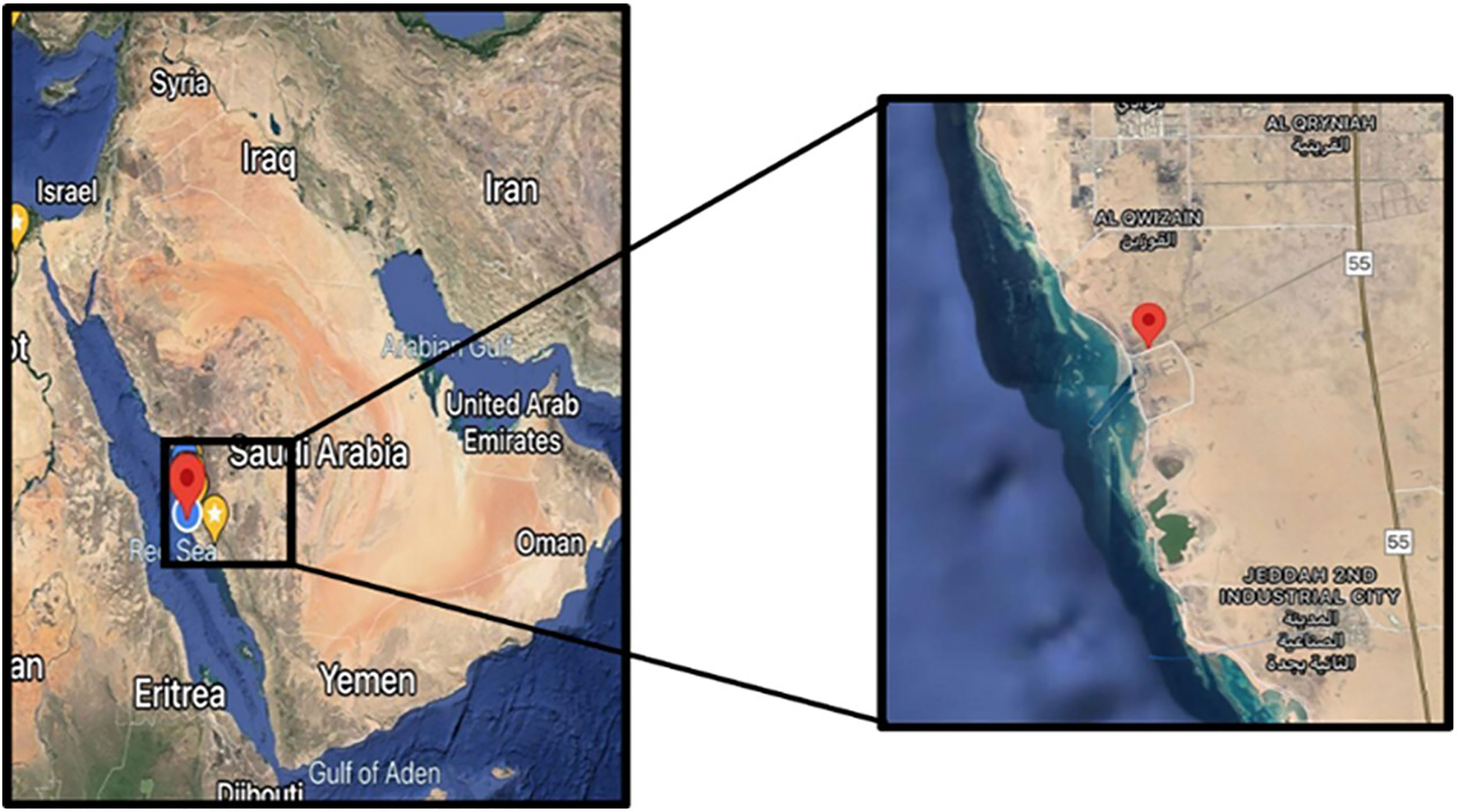
Soil samples were collected from Southern corniche (Jeddah, Saudi Arabia), which is a saline sandy area where very few plants grow (GPS: 21.2181995–39.1756291). Sc samples were collected from crust soil of S. monoica, Sr samples were collected from rhizosphere soil of S. monoica, Dc samples were collected from crust soil of D. glaucum, and Dr samples were collected from rhizosphere soil of D. glaucum. A control sample (c) was also collected from soil samples where no plant growth was observed (Figure 1).
2.2 DNA Extraction
FastDNA™ Spin Soil Kit (MP Biomedicals, Santa Ana, CA, USA) was used to extract total genomic DNA for the next-generation sequencing (NGS) according to the manufacturer’s instructions (Furtak et al., 2019). The DNA was visualized on 1% TAE agarose gel to assess DNA quality. DNA quality was measured using nanodrop (Implen NanoPhotometer® N60/N50) and fluorometric quantification using iQuant™ Broad Range dsDNA Quantification Kit. Extracted gDNA was diluted in sterile water to 10 ng µl−1 and stored at −20°C until further processing.
2.3 PCR Amplification and Next-Generation Sequencing
The purified gDNA were proceeded with NGS by amplifying the hypervariable V3–V4 region of the 16S rRNA gene using 341F (5′CCTACGGGNGGCWGCAG3′) and 785R (5′GACTACHVGGGTATCTAATCC3′) primers (Furtak et al., 2019). PCRs were carried out with REDiant 2× PCR Master Mix (1st BASE, Kuala Lumpur, Malaysia). Library construction was carried out in 2 steps where selected regions (16S V3–V4) were amplified using locus-specific sequence primers with forward (5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG) and reverse (5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG) overhang adapters. PCRs were carried out with KOD-Multi & Epi-® (Toyobo, Osaka, Japan) (1st BASE). In the second step, dual indices were attached to the amplicon PCR using Illumina (San Diego, CA, USA) Nextera XT Index Kit v2 following the manufacturer’s protocols. The quality of the libraries was measured using Agilent (Santa Clara, CA, USA) Bioanalyzer 2100 System by Agilent DNA 1000 Kit and fluorometric quantification by Helixyte Green™ Quantifying Reagent. The libraries were normalized and pooled according to Illumina-suggested protocols, and sequencing was done using MiSeq platform using 300 PE (1st BASE). Primers and adapters were removed followed by the merging of forward and reverse reads. Sequences shorter than 150 bp were removed, and the reads were made even before the data were proceeded with operational taxonomic unit (OTU) clustering using SILVA, GreenGenes, RDP, UNITE, and other databases for taxonomic assignments.
2.4 Statistical Analysis
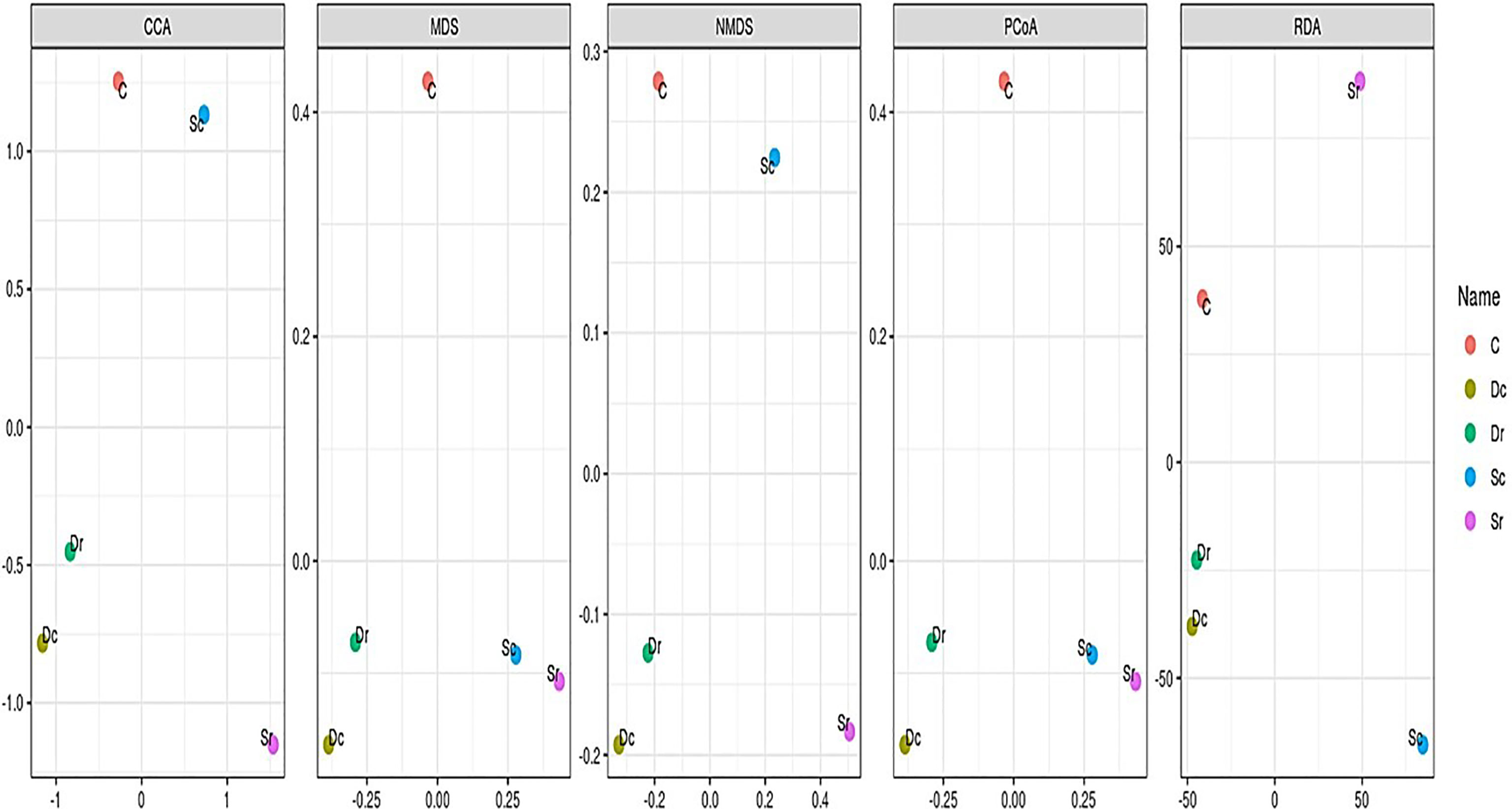
One-way ANOVA was used for multiple comparisons of the 5 samples. For α-diversity, Chao1, Shannon, Simpson, InvSimpson, and Fisher diversity indices were used to estimate richness and diversity. Rarefaction curves were also generated based on the average number of observed OTUs to compare the relative levels of OTU diversity. For β-diversity, UniFrac distant matrix, principal coordinates analysis (PCoA), multidimensional scaling/non-metric multidimensional scaling (MDS/NMDS), canonical correspondence analysis/redundancy analysis (CCA/RDA), and UPGMA-Tree were used. PCoA plots were prepared based on weighted and unweighted UniFrac distance metrics to observe the similarities between soil samples and the clustering of the different soil groups. Multivariate parametric analysis (DESeq) was used to analyze parametric data, while analysis of similarity (ANOSIM) was used to analyze the non-parametric data. Venn diagram and double dendrogram clustered heatmaps were generated to visualize the data and observe how the soil samples will cluster.
2.5 Prediction of Bacterial Community Function
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict functional composition in the metagenome of the 5 soil samples generated using the GreenGenes database (Wu et al., 2019; Yurgel et al., 2019). PICRUSt metagenome inferences were carried out based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (KO) and Enzyme Commission numbers (EC numbers). A dendrogram with a heatmap was generated to visualize the data and observe how the soil samples will cluster.
3 Results
3.1Composition
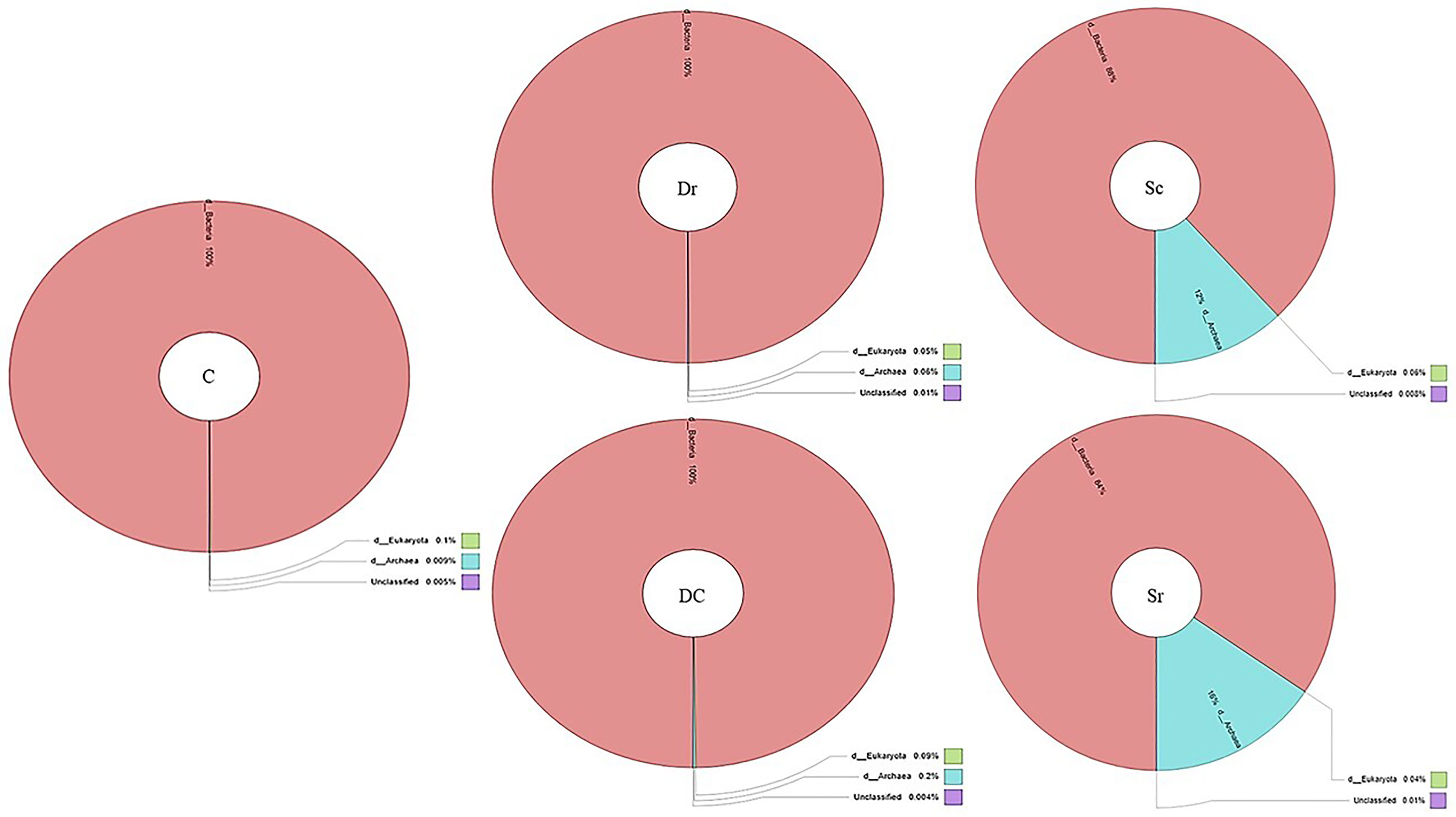
The 16s V3–V4 region analysis detected archaeal strains in Sc and Sr samples with an abundance of 12% and 16%, respectively, while other samples (C, Dc, and Dr) contained 100% bacterial strains (Figure 2). A total of 19–23 phyla were found in the studied soil samples with diverse compositions, which showed the richness of the soil samples and interactions with plants. Using 1% as a threshold, a total of 19 phyla, 34 classes, and 76 orders were detected, which showed the richness of the soil microbiome. Top 10 composition analysis of soil samples was carried out, and the results showed an interaction between soil microbiome and plants (Supplementary Figure 1). For instance, the composition of the control sample was relatively even among typical soil bacteria, while samples associated with plants showed diverse distribution and the rise of new phyla, namely, Euryarchaeota (Halobacterota).
The top 10 phyla composition of the soil samples was Proteobacteria (23% ± 6%), Actinobacteria (19% ± 12%), Firmicutes (19% ± 8%), Gemmatimonadota (9% ± 4%), Bacteroidota (6% ± 4%), Halobacterota (6% ± 8%), Cyanobacteria (6% ± 6%), Cloroflexi (5% ± 3%), Planctomycetota (3% ± 2%), and Myxococcota (3% ± 2%). The highest variation between soil samples was in halophilic archaea Halobacterota and Cyanobacteria composition, while Proteobacteria percentage was the most similar. Halobacterota, which is from archaea, was only present in S. monoica-associated soil samples (Sc and Sr), while Cyanobacteria was almost absent in D. glaucum samples (Dc and Dr). Myxococcota was also mainly present in plant-associated soil samples (Dc, Dr, S, c, and Sr).
The top 10 class composition of the soil samples was Bacilli (22% ± 9%), Alphaproteobacteria (17% ± 5%), Actinobacteria (17% ± 11%), Gammaproteobacteria (12% ± 6%), Longimicrobia (8% ± 4%), Halobacteria (7% ± 9%), Cyanobacteria (6% ± 8%), Acidimicrobiia (5% ± 5%), Bacteroidia (4% ± 3%), and Planctomycetes (3% ± 2%). The highest variation was observed in Halobacteria and Cyanobacteria classes, while the highest similarity was observed in the Alphaproteobacteria class. All the top 10 classes showed high variations with a coefficient of variation (CV) of more than 30%.
Order composition showed a similar trend to phyla and class compositions where the highest variation was observed in Halobacteriales and Cyanobacteriales orders. The top 10 order composition of the soil samples was Bacillales (28% ± 13%), Longimicrobiales (12% ± 6%), Frankiales (10% ± 8%), Halobacteriales (9% ± 14%), Rhizobiales (9% ± 6%), Burkholderiales (9% ± 8%), Paenibacillales (7% ± 2%), Cytophagales (6% ± 4%), Rhodobacterales (5% ± 3%), and Cyanobacteriales (5% ± 7%). The highest similarity between soil samples was observed in the Paenibacillales order.
The top 10 family compositions showed high variations (CV% > 30) in 9 out of 10 families. The highest variations were observed in Micrococcaceae, Balneolaceae, and Nostocaceae. Other differences observed were that Sr samples contained almost twice Bacillaceae as Sc. The top 10 family composition was Bacillaceae (33% ± 15%), Longimicrobiaceae (15% ± 8%), Geodermatophilaceae (10% ± 8%), Paenibacillaceae (8% ± 1%), Sphingomonadaceae (7% ± 5%), Rhodobacteraceae (7% ± 4%), Beijerinckiaceae (7% ± 4%), Micrococcaceae (6% ± 6%), Balneolaceae (4% ± 9%), and Nostocaceae (4% ± 7%). The highest similarity between soil samples was observed in the Paenibacillaceae family.
The top 10 genera composition also showed high variations (CV% > 30) in 9 out of 10 genera. The highest variation was observed in the Sphaerospermopsis genus, which was mainly present in Sc and absent in Dc and Dr. Pontibacter was mainly present in the control sample and absent in Sr, while Halomonas was mainly present in Sr and absent in Dc and Dr. Geodermatophilus genus was absent in Sr and present at low abundance (1%) in Sc. The top 10 genera composition of the soil samples was Bacillus (36% ± 14%), Longimicrobiaceae (19% ± 11%), Geodermatophilus (8% ± 7%), Microvirga (7% ± 5%), Ammoniphilus (7% ± 2%), Rubellimicrobium (6% ± 5%), Blastococcus (5% ± 3%), Halomonas (5% ± 10%), Pontibacter (4% ± 5%), and Sphaerospermopsis (3% ± 6%). The highest similarity between soil samples was observed in the Ammoniphilus genus.
3.2 Clustering
3.2.1 Operational Taxonomic Units and Diversity Indices
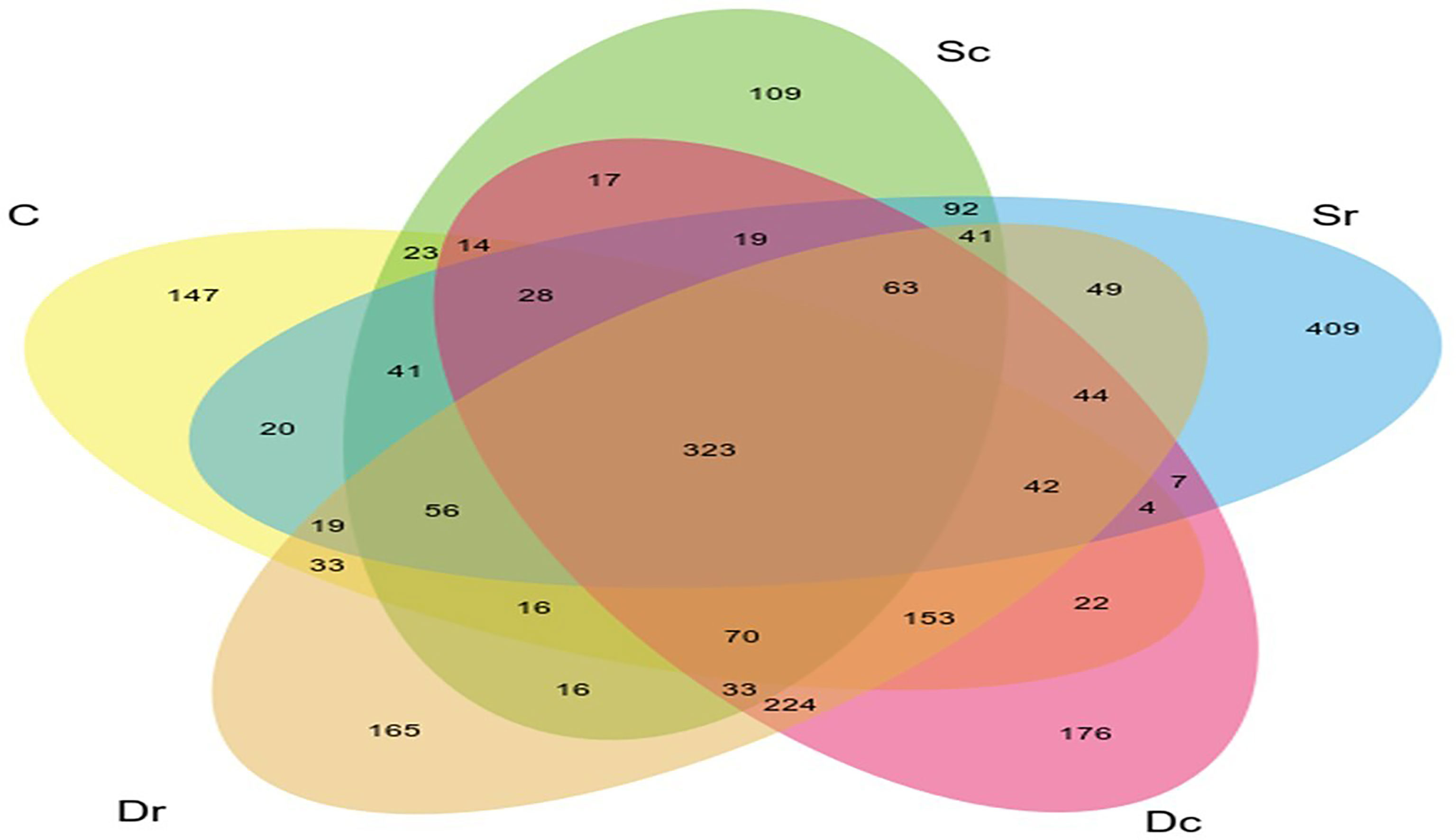
Figure 3 highlights the relationships, overlaps, and differences between studied soil samples (C, Dc, Dr, Sc, and Sr).
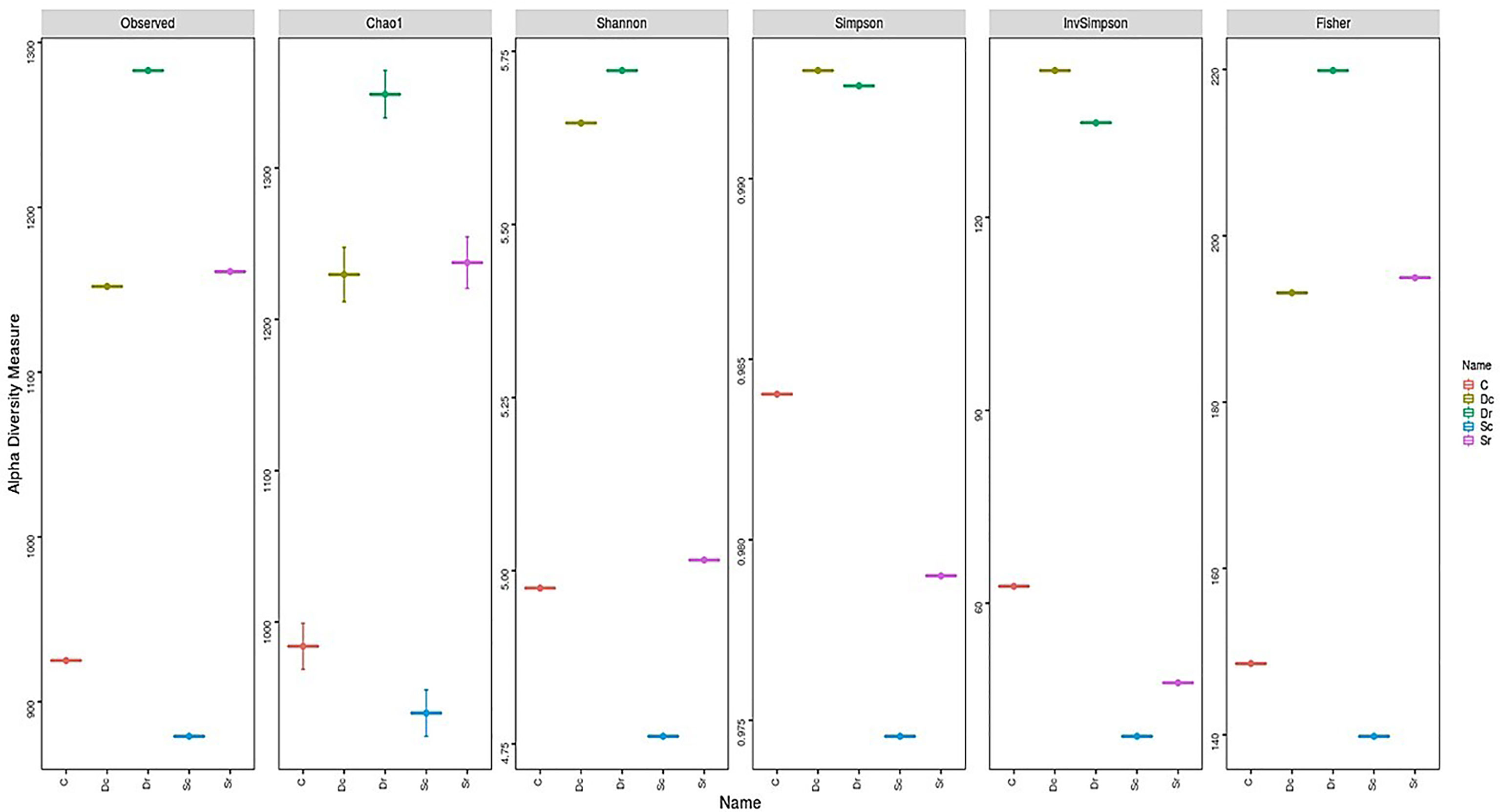
Sc had less OTUs and lower diversity as compared to the control sample (C), while Dr showed the highest number of OTUs and diversity. All α-diversity indices determined Sc as the sample with the lowest diversity and richness (Figure 4). However, the ranking for other samples varied across different indices. Sr and Dc had similar OTUs but showed different results in α-diversity indices. Most of the variation in α-diversity indices was in Simpson and InvSimpson indices. For instance, Dr was the richest sample in all indices except in Simpson and InvSimpson. The highest variations across all indices were observed in Sr, which ranged from being ranked 4th in diversity (Simpson and InvSimpson indices) to being ranked 2nd (Chao1 and Fisher). D. glaucum samples showed higher diversity than S. monoica at the family level, and Balneolaceae and Nostocaceae were only present in S. monoica; however, they were present in small amounts. For example, in the top 10 families, Balneolaceae and Nostocaceae made up 16% and 20% in Sr and Sc, respectively; however, they only made up <10% of all the families detected in S. monoica samples. In terms of sample type, rhizosphere-related samples showed higher diversity than crust-related samples.
In β-diversity, several analysis methods (CCA, RDA, NMDS, MDS, and PCoA) were carried out to compare the 5 samples (Figure 5). The control sample (C) was often clustered separately, while Dc and Dr were always clustered together. Sample Sc and Sr clustered differently between different methods. NMDS and RDA showed the highest sensitivity and clustered the 5 samples into 4 clusters, while PCoA and MDS produced the most accurate clusters by showing 3 clusters (C, Dc and Dr, and Sc and Sr).
3.2.2 Heatmaps and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States Analysis
Heatmap-based clustering of S. monoica, D. glaucum, and control samples showed that at phylum, class, order, and family levels, all 3 samples clustered separately, with D. glaucum being closer to control than S. monoica. At genus and species levels, the rhizosphere sample of S. monoica (Sr) became a unique cluster, while the crust of S. monoica (Sc) clustered with the control sample. Aside from numerous unidentified or uncultured species, this unique Sr cluster was due to the dominance of halophilic bacteria that belong to Halomonas and Halofilum genera (Supplementary Figure 2).
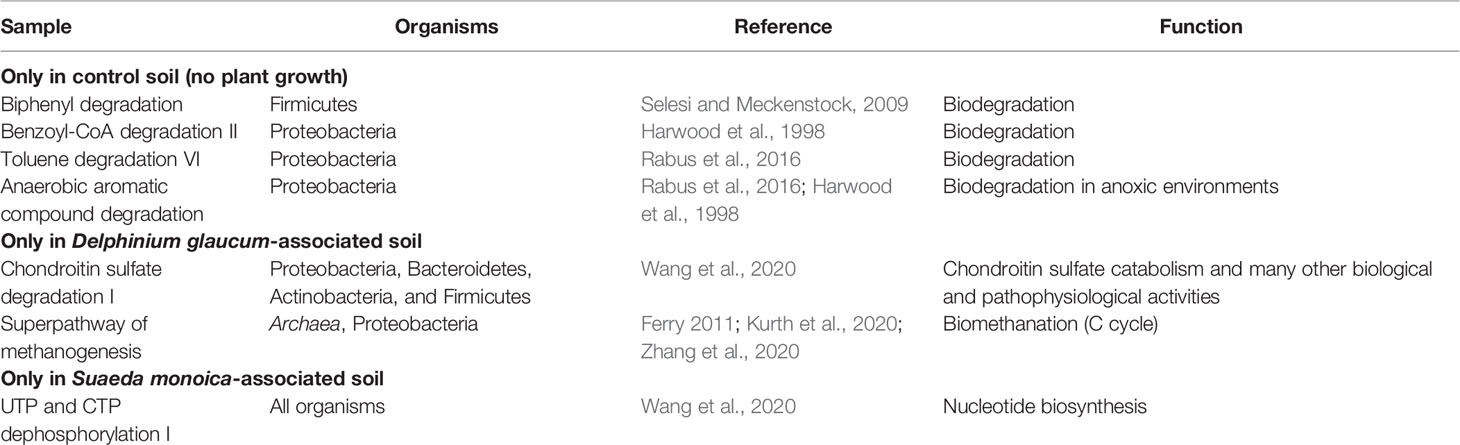
EC- and KO-based analyses of functional genes and pathways showed that S. monoica, D. glaucum, and control samples all clustered separately, which showed the significant difference between different groups of samples. The results also showed that 5 pathways were specific to control, 11 pathways were observed only in D. glaucum samples, 12 pathways were expressed in S. monoica samples only, and 9 pathways were common in all samples. Interestingly, 19 pathways were mainly observed in D. glaucum, while a total of 37 pathways were observed mainly in S. monoica. Table 1 summarizes the unique pathways detected in various samples and their functions, while Table 2 summarizes the pathways that were upregulated in certain samples and their functions (Supplementary Figure 3).
4 Discussion
Proteobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Acidobacteria, and Firmicutes are dominant phyla in soil (Dube et al., 2019; Wolińska, 2019). The results of this study were similar, as the top 5 phyla detected in soil samples were Proteobacteria (23% ± 6%), Actinobacteriota (19% ± 12%), Firmicutes (19% ± 8%), Gemmatimonadota (9% ± 4%), and Bacteroidota (6% ± 4%). Gemmatimonadota was present in high concentrations and was previously established as a typical soil phylum (Lena and Suda, 2018). Proteobacteria phylum and Paenibacillaceae belonging to Firmicutes phylum were the most common in all soil samples. Proteobacteria and Firmicutes are known to be dominant phyla in soil (Dube et al., 2019). This can be observed in the control samples where aside from the overall evenness of the phyla’s distribution, the percentages of Proteobacteria, Actinobacteria, and Firmicutes were similar, while soil samples associated with plants showed unevenness. In this study, the top 10 phylum analysis of S. monoica showed that both Proteobacteria and Actinobacteriota were low, which could be due to the abundance of halophilic archaea belonging to Halobacterota phylum (14.2% ± 1.4). Archaea contains the conserved region 16s V3–V4 as reported by recent studies (Lena & Suda, 2018; McGovern et al., 2018; Bukin et al., 2019; Willis et al., 2019). These halophilic methanogenic archaea are known to be common in soil; however, they are less tolerant to desiccation and oxygen exposure (Conrad, 2020). This can be supported by the fact Halobacterota content in the rhizosphere samples, which are more protected from heat and oxygen, was higher than in crust samples. In addition, the detection of archaea in the crust and rhizosphere of S. monoica samples only showed that soil microbes respond to differences in soil chemistry, which was in line with Dube et al. (2019), who studied the effect of long-term agriculture activities on the bacterial diversity in the soil.
Previous research also suggested that bacteria in the soil and their plant host interact differently in response to the same stress (e.g., salinity) (Kearl et al., 2019). Other studies also reported that plants excrete certain chemicals to recruit certain soil bacteria or suppress others to increase their resistance to diseases (Berendsen et al., 2018; Liu and Brettell, 2019). Myxococcota has been recently proposed as a new phylum and was mainly present in plant-associated soil samples (Dc, Dr, Sc, and Sr). The phylum was proposed due to the unique “pack hunting” predatory strategy of Myxococcota in which they feed on other bacteria by swarming and secreting lytic enzymes (Waite et al., 2020). The results also showed that when Chloroflexi was high in D. glaucum, cyanobacteria were low. Chloroflexi is considered light-harvesting (Thweatt et al., 2019) and was previously found to outcompete cyanobacteria in nitrogen fixation (Lacap et al., 2011) and is capable of carbon fixation (Ward et al., 2018). Rhizosphere of halophytes was reported to have diverse bacterial communities that promote plant growth by increasing the uptake of organic and minerals from the soil (Mehnaz et al., 2017; Kearl et al., 2019). In this study, rhizosphere-related samples showed slightly higher diversity than crust-related samples. The average number of phyla was 13 and 14 in rhizosphere and crust samples, respectively. The average number of classes was 38 and 36 in rhizosphere and crust samples, respectively. Overall, the maximum identified difference was observed at the genus level where S. monoica’s crust was high in Sphaerospermopsis, rhizosphere of S. monoica was high in Halomonas and Bacillus, and control was high in Pontibacter. Halophilic bacteria stimulate plant growth via many mechanisms such as increasing photosynthesis, reducing sodium uptake from the soil, and creating biofilms to trap water and nutrients (Kearl et al., 2019). In this study, the rhizosphere of S. monoica contained Halomonas genus, which was previously found to be common in the root and rhizosphere of halophytes and stimulated the growth of plants such as alfalfa in high-salinity conditions (Kearl et al., 2019). A recent study also reported that high salinity significantly reduced Actinobacteria (−0.99) and increased Firmicutes (0.88) (Kuznetsova et al., 2020). This can be supported by the fact that in D. glaucum, where Firmicutes was low, Actinobacteria was high. In terms of Sphaerospermopsis abundance in S. monoica’s crust, Cyanobacteria from Sphaerospermopsis BCCUSP55 genus could be related to nitrogen fixation at the soil surface. Sphaerospermopsis genus was previously reported to be closely related to a well-known nitrogen-fixing cyanobiont bacteria known as Nostoc (Gunawardana, 2020). This can be supported by the fact that at the family level, Nostocaceae cyanobacteria were abundant in S. monoica-associated samples, especially S. monoica’s crust, while Balneolaceae was abundant in Sr. Another species, Sphaerospermopsis aphanizomenoides, was reported to not only grow faster than other Nostocales families in high temperature but also to be correlated with phytoplankton (Budzyńska et al., 2019).
V3–V4 region was found to produce high richness and diversity (García-López et al., 2020). In this study, variations at the species level were high; however, it was mostly unidentified or uncultured species, which indicated that the V3–V4 region is efficient for higher-order classification. A similar issue was encountered in this study where V3–V4 region results of genus composition of D. glaucum also contained up to 3% unidentified Sphingomonas spp. This genus is well studied, and the ambiguity of these species could be due to the V3–V4 region limitation as the genus that we studied. Rubellimicrobium (7%) and Kocuria (6%) are also well-studied, yet their species were not determined. A previous study also reported that the Rubellimicrobium genus completely disappeared once plants grew on the soil (Köberl et al., 2013), despite Kocuria species being known to tolerate salinity, promote plant growth, and increase their ability to resist disease (Goswami et al., 2014). Recent studies recommended accurate species classification in other regions such as V2–V3 (Bukin et al., 2019) or other newer methods such as sFL16S (Jeong et al., 2021). However, some of the observed unidentified species could be due to the novelty of the species, as many belonged to relatively new genera and families. For instance, the rhizosphere of S. monoica contained a relatively high amount of unidentified Aliifodinibius spp. belonging to the Balneolaceae family. Aliifodinibius genus was described in 2013 (Wang et al., 2013) and currently contains only 6 known species, namely, Aliifodinibius roseus (identified 2013), Aliifodinibius sediminis (identified 2013), Aliifodinibius halophilus (identified 2016), Aliifodinibius salicampi (identified 2017), Aliifodinibius salipaludis (identified 2020), and Aliifodinibius saliphilus (identified 2020) (Zhao et al., 2020). The genus composition of D. glaucum also contained up to 3% of a relatively new genus called Lacunisphaera, which was described in 2017 (Rast et al., 2017). New genera under the same family (Opitutaceae) have also been proposed as recently as 2018 (Rochman et al., 2018). Other highly unclassified bacteria were observed mainly in the control sample, which was Pontibacter (7%). Pontibacter genus is relatively new, described in 2005, and novel species are being added as recently as Pontibacter oryzae and Pontibacter chitinilyticus in 2019 (Chhetri et al., 2019a; Chhetri et al., 2019b), while Pontibacter pudoricolor and Pontibacter russatus were described in 2020 (Maeng et al., 2020). Hence, novel halophilic Aliifodinibius spp. and Lacunisphaera spp. could be isolated from the sample or site. Pontibacter genus was recently found to be extremely tolerant to physiochemical and environmental stresses (Belov et al., 2019).
In terms of OTUs and diversity indices, the rhizosphere of S. monoica showed the highest variations across different indices ranging from being ranked 4th in diversity (Simpson and InvSimpson indices) to being ranked 2nd (Chao1 and Fisher). Most variations in ranking can be attributed to Simpson and InvSimpson in terms of indices and rhizosphere of S. monoica in terms of samples. Diversity indices measure and are influenced by richness, commonly measured by the Shannon index, and evenness, commonly measured by the Simpson index (MacDonald et al., 2017). An inverse relationship between species richness and evenness was also reported previously where detection of rare/less abundant species decreased evenness and increases richness (MacDonald et al., 2017). In this study, OTU numbers correlated with richness, Chao1, and Fisher indices only and not Shannon, while Simpson and InvSimpson indices gave similar results to Pielou’s evenness. Additionally, the rhizosphere soil of D. glaucum was the richest except in Simpson, InvSimpson, and Pielou’s evenness, while the rhizosphere of S. monoica’s and D. glaucum’s crust has similar OTUs but showed different results in α-diversity indices. These 2 samples (Sr and Dc) had significantly different composition at phylum, order, and family levels, which showed that OTUs alone is not a good indicator of diversity. This complexity and varying rankings of studied samples have been previously addressed and linked with varying richness (total number of species), the proportion of each species between different samples, detection of rare species, and other statistical differences between the indices (MacDonald et al., 2017; Daly et al., 2018). Considering that majority of the species were not identified, the Simpson index could be more accurate because they are less sensitive to minor/rare species (Morris et al., 2014); hence, D. glaucum samples showed higher diversity than S. monoica. Additionally, heatmap results showed that S. monoica, D. glaucum, and control samples showed that at phylum, class, order, and family levels, all 3 samples clustered separately. This is also supported by ordination analysis by PCoA that clustered the 5 samples into 3 clusters representing the 3 main soil samples (control, D. glaucum, and S. monoica).
EC- and KO-based analyses of functional genes and pathways showed that S. monoica, D. glaucum, and control samples all clustered separately, which showed the significant difference between different groups of samples. However, the differences in the pathways could be due to the difference in species compositions. For example, control samples that have the least species diversity showed the least unique pathways (n = 5), while D. glaucum samples and S. monoica samples showed more than 10 unique pathways each. This was expected, as the microbiome composition of D. glaucum samples and S. monoica samples contained archaeal species. Table 1 shows the unique pathway predicted in the studied samples. Control samples expressed mainly aromatic compound degradation pathways such as biphenyl degradation, benzoyl-CoA degradation, and toluene degradation, which free up nutrients (Harwood et al., 1998; Rabus et al., 2016; Selesi and Meckenstock, 2009). On the contrary, S. monoica appeared to prefer nucleoside and nucleotide degradation pathways as sources of nutrients and energy (Wang et al., 2020). Interestingly, the chondroitin sulfate degradation I pathway, which is produced to degrade animal carcass tissues to provide nutrients for the plant, was predicted in D. glaucum, which is known to be extremely poisonous to mammals, causing paralysis in cattle (Manners et al., 1998).
Table 2 summarizes the pathways predicted to be upregulated in the studied samples. Control samples showed the upregulation of very few stress-related pathways such as superpathway of polyamine biosynthesis III, which is linked with spermidine synthesis. Spermidine is produced by Proteobacteria as a stress tolerance mechanism (Barbagallo et al., 2011). Unlike control samples, plant-associated soil samples expressed numerous plant-promoting trains such as the acquisition of nitrogen, phosphorus, and essential minerals (e.g., vitamins) and the production of antibiotics to combat pathogens (Ahemad and Kibret, 2014). For example, D. glaucum samples showed increased expression of superpathway of mycolyl–arabinogalactan–peptidoglycan complex biosynthesis and neopentalenoketolactone and pentalenate biosynthesis, which could be due to the abundance of Actinobacteria order (Crick and Brennan, 2008). Superpathway of mycolyl–arabinogalactan–peptidoglycan complex biosynthesis is also a cell wall biosynthesis. This indicated a bacterial growth promotion, which is supported by the high diversity observed in D. glaucum. In terms of S. monoica, the predicted pathways were the upregulation of l-rhamnose degradation, glucose degradation, and nitrate reduction, which could be due to the abundance of halotolerant strains such as Halomonas elongata. Previous studies reported that Halomonas strains contain high quantities of glucose, mannose, and rhamnose and express nitrate reductases (Maheshwari and Saraf, 2016; Joulak et al., 2021). Moreover, several lipid biosynthesis pathways were upregulated such as archaetidylinositol and CDP-archaeol biosynthesis pathways. Previous studies also linked the abundance of lipids with the presence of archaea (Maheshwari and Saraf, 2016; Caforio and Driessen, 2017). The major differences between soil samples could be explained by the associated plant species; however, a number of differences in terms of predicted pathways were also observed. The main pattern observed in rhizosphere samples was the expression of sugar synthesis and degradation pathways. For example, l-rhamnose, lactose, galactose, and glucose degradation pathways were predicted in S. monoica rhizosphere. The high expression of sugar synthesis and degradation pathways was expected, as the rhizosphere is known to be a sugar-rich habitat that promotes the growth of the associated microbiome (Jha and Subramanian, 2018). Taken together, predicted expression pathways were correlated with the species composition, as all samples were collected from geographically adjacent sites.
Conclusion
The most common phyla in soil samples were Proteobacteria, Actinobacteriota, Firmicutes, Gemmatimonadota, and Bacteroidota, which is a typical bacterial community found in the soil microbiome. The control sample that was taken from the soil with no plant growth showed a relatively even distribution of major phyla, while plant-associated samples showed a significant increase in certain phyla as well as the growth of archaea. The V3–V4 region analysis gave a good background on the general and higher-order classification. It successfully clustered the 5 samples into 3 clusters (control, D. glaucum, and S. monoica) at phylum, class, order, and family levels. In terms of genus and species levels, future research can be carried out on amplifying the V2–V3 region to get a more accurate lower classification. The soil composition of S. monoica-associated samples contained relatively high amounts of halophilic archaea such as Salarchaeum japonicum and Halococcus hamelinensis, halophilic bacteria such as Halomonas elongata, and unidentified cyanobacteria such as Sphaerospermopsis spp. This could explain why S. monoica had lower diversity than D. glaucum and control samples. The soil composition of D. glaucum contained up to 3% each for Lacunisphaera spp. and Sphingomonas spp. Few relatively novel genera were detected in high abundances such as Aliifodinibius, Pontibacter, and Lacunisphaera. This indicated that novel species could be isolated from the soil samples and used for future research. Predicted pathways indicated that most of the differences between soil samples were due to the associated plant species and microbiome composition.
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA821426, PRJNA821365 and PRJNA821368.
Author Contributions
RJ, RA and NasB: Data collection and project administration, HS: Statistical analyses and original draft writing, MAl: Project administration and original draft writing, AS: Funding acquisition, AA and MR: Software and formal analysis, LB, AF and MAr: Final draft writing, NabB: Conceptualization and Design of the study, MB: Design of the study and activities supervision. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This project was funded by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R31) and Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R31), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.865834/full#supplementary-material
References
Abbanat D. R., Ferry J. G. (1990). Synthesis of Acetyl Coenzyme A by Carbon Monoxide Dehydrogenase Complex From Acetate-Grown Methanosarcina Thermophila. J. Bacteriol. 172 (12), 7145–7150. doi: 10.1128/jb.172.12.7145-7150.1990
Ahemad M., Kibret M. (2014). Mechanisms and Applications of Plant Growth Promoting Rhizobacteria: Current Perspective. J. King. Saud. Univ-Sci. 26 (1), 1–20. doi: 10.1016/j.jksus.2013.05.001
Ahmad M., Anjum N. A., Asif A., Ahmad A. (2020). Real-Time Monitoring of Glutathione in Living Cells Using Genetically Encoded FRET-Based Ratiometric Nanosensor. Sci. Rep. 10 (1), 992. doi: 10.1038/s41598-020-57654-y
Alderwick L. J., Seidel M., Sahm H., Besra G. S., Eggeling L. (2006). Identification of a Novel Arabinofuranosyltransferase (AftA) Involved in Cell Wall Arabinan Biosynthesis in Mycobacterium Tuberculosis. J. Biol. Chem. 281 (23), 15653–15661. doi: 10.1074/jbc.M600045200
Al-Said M. S., Siddiqui N. A., Mukhair M. A., Parvez M. K., Alam P., Ali M., et al, et al. (2017). A Novel Monocyclic Triterpenoid and a Norsesquaterpenol From the Aerial Parts of Suaeda Monoica Forssk. Ex JF Gmel With Cell Proliferative Potential. Saudi Pharm. J. 25, 1005–1010. doi: 10.1016/j.jsps.2017.03.008
Altwaty N. H., El-Sayed O. E., Aly N. A., Baeshen M. N., Baeshen N. A. (2016). Molecular and cytogenetic assessment of dipterygium glaucum genotoxicity. Anais da Academia Bras. Cienc. 88, 623–634. doi: 10.1590/0001-3765201620150208
Alzahrani D., Albokhari E., Yaradua S., Abba A. (2020). Complete plastome genome of dipterygium glaucum, dipterygieae, cleomaceae. Mitochondrial DNA Part B 5 (2), 1872–1873. doi: 10.1080/23802359.2020.1750978
Aoshima M., Igarashi Y. (2006). A Novel Oxalosuccinate-Forming Enzyme Involved in the Reductive Carboxylation of 2-Oxoglutarate in Hydrogenobacter Thermophilus TK-6. Mol. Microbiol. 62 (3), 748–759. doi: 10.1111/j.1365-2958.2006.05399.x
Ayyappan D., Balakrishnan V., Ravindran K. (2013). Potentiality of Suaeda Monoica Forsk. A Salt Marsh Halophyte on Restoration of Saline Agricultural Soil. World Appl. Sci. J. 28, 2026–2032. doi: 10.5829/idosi.wasj.2013.28.12.2136
Azami Y., Hattori A., Nishimura H., Kawaide H., Yoshimura T., Hemmi H. (2014). (R)-Mevalonate 3-Phosphate Is an Intermediate of the Mevalonate Pathway in Thermoplasma Acidophilum. J. Biol. Chem. 289 (23), 15957–15967. doi: 10.1074/jbc.M114.562686
Bærentsen R. L., Brodersen D. E., Zhang Y. E. (2019). Evolution of the Bacterial Nucleosidase PpnN and Its Relation to the Stringent Response. Microbial Cell (Graz Austria) 6 (9), 450–453. doi: 10.15698/mic2019.09.692
Barbagallo M., Di Martino M. L., Marcocci L., Pietrangeli P., De Carolis E., Casalino M., et al. (2011). A New Piece of the Shigella Pathogenicity Puzzle: Spermidine Accumulationby Silencing of the speG Gene. PloS One 6 (11), e27226. doi: 10.1371/journal.pone.0027226
Bashiri G., Antoney J., Jirgis E. N. M., Shah M. V., Ney B., Copp J., et al. (2019). A Revised Biosynthetic Pathway for the Cofactor F420 in Prokaryotes. Nat. Commun. 10 (1), 1558. doi: 10.1038/s41467-019-09534-x
Batanouny K. H., Baeshin N. A. (1982). Studies on the flora of Arabia, II. The Medina-badr road Saudi Arabia. Bull. Fac. Sci. 6, 1–26.
Batanouny K. H., Baeshin N. A. (1983). Plant communities along the Medina-badr road across the hejaz mountains, Saudi Arabia. Vegetatio 53 (1), 33–43. doi: 10.1007/BF00039769
Basu A., Phale P. S. (2006). Inducible Uptake and Metabolism of Glucose by the Phosphorylative Pathway in Pseudomonas Putida CSV86. FEMS Microbiol. Lett. 259 (2), 311–316. doi: 10.1111/j.1574-6968.2006.00285.x
Bellais S., Arthur M., Dubost L., Hugonnet J.-E., Gutmann L., van Heijenoort J., et al. (2006). Aslfm, the D-Aspartate Ligase Responsible for the Addition of D-Aspartic Acid Onto the Peptidoglycan Precursor of Enterococcus Faecium. J. Biol. Chem. 281 (17), 11586–11594. doi: 10.1074/jbc.M600114200
Belov A. A., Cheptsov V. S., Vorobyova E. A., Manucharova N. A., Ezhelev Z. S. (2019). Stress-Tolerance and Taxonomy of Culturable Bacterial Communities Isolated From a Central Mojave Desert Soil Sample. Geosciences 9 (4), 166. doi: 10.3390/geosciences9040166
Berendsen R. L., Vismans G., Yu K., Song Y., de Jonge R., Burgman W. P., et al. (2018). Disease-Induced Assemblage of a Plant-Beneficial Bacterial Consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Bhat B. A., Mir W. R., Sheikh B. A., Rather M. A., Mir M. A. (2022). In Vitro and in Silico Evaluation of Antimicrobial Properties of Delphinium Cashmerianum L., a Medicinal Herb Growing in Kashmir, India. J. Ethnopharmacol. 291, 115046. doi: 10.1016/j.jep.2022.115046
Blank D., Wolf L., Ackermann M., Silander O. K. (2014). The Predictability of Molecular Evolution During Functional Innovation. Proc. Natl. Acad. Sci. U. S. A. 111 (8), 3044–3049. doi: 10.1073/pnas.1318797111
Bologna F. P., Campos-Bermudez V. A., Saavedra D. D., Andreo C. S., Drincovich M. F. (2010). Characterization of Escherichia Coli EutD: A Phosphotransacetylase of the Ethanolamine Operon. J. Microbiol. (Seoul Korea) 48 (5), 629–636. doi: 10.1007/s12275-010-0091-0
Brouns S. J. J., Walther J., Snijders A. P. L., van de Werken H. J. G., Willemen H. L. D. M., Worm P., et al. (2006). Identification of the Missing Links in Prokaryotic Pentose Oxidation Pathways: Evidence for Enzyme Recruitment. J. Biol. Chem. 281 (37), 27378–27388. doi: 10.1074/jbc.M605549200
Budzyńska A., Rosińska J., Pełechata A., Toporowska M., Napiórkowska-Krzebietke A., Kozak A., et al. (2019). Environmental Factors Driving the Occurrence of the Invasive Cyanobacterium Sphaerospermopsis Aphanizomenoides (Nostocales) in Temperate Lakes. Sci. Total Environ. 650, 1338–1347. doi: 10.1016/j.scitotenv.2018.09.144
Bukin Y. S., Galachyants Y. P., Morozov I., Bukin S., Zakharenko A., Zemskaya T. (2019). The Effect of 16S rRNA Region Choice on Bacterial Community Metabarcoding Results. Sci. Data 6, 1–14. doi: 10.1038/sdata.2019.7
Caforio A., Driessen A. J. (2017). Archaeal Phospholipids: Structural Properties and Biosynthesis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1862 (11), 1325–1339. doi: 10.1016/j.bbalip.2016.12.006
Campbell M. A., Nyerges G., Kozlowski J. A., Poret-Peterson A. T., Stein L. Y., Klotz M. G. (2011). Model of the Molecular Basis for Hydroxylamine Oxidation and Nitrous Oxide Production in Methanotrophic Bacteria. FEMS Microbiol. Lett. 322 (1), 82–89. doi: 10.1111/j.1574-6968.2011.02340.x
Chhetri G., Kim J., Kim H., Kim I., Seo T. (2019a). Pontibacter Oryzae Sp. Nov., a Carotenoid-Producing Species Isolated From a Rice Paddy Field. Antonie Van Leeuwenhoek 112, 1705–1713. doi: 10.1007/s10482-019-01298-0
Chhetri G., Kim J., Kim I., Kim M. K., Seo T. (2019b). Pontibacter Chitinilyticus Sp. Nov., a Novel Chitin-Hydrolysing Bacterium Isolated From Soil. Antonie Van Leeuwenhoek 112, 1011–1018. doi: 10.1007/s10482-019-01235-1
Conrad R. (2020). Methane Production in Soil Environments—Anaerobic Biogeochemistry and Microbial Life Between Flooding and Desiccation. Microorganisms 8, 881. doi: 10.3390/microorganisms8060881
Cook A. M., Denger K. (2002). Dissimilation of the C2 Sulfonates. Arch. Microbiol. 179 (1), 1–6. doi: 10.1007/s00203-002-0497-0
Crick D. C., Brennan P. J. (2008). Biosynthesis of the Arabinogalactan‐Peptidoglycan Complex of Mycobacterium Tuberculosiss. Mycobacterial Cell Envelope, 25–40. doi: 10.1093/glycob/11.9.107r.
Daly A. J., Baetens J. M., De Baets B. (2018). Ecological Diversity: Measuring the Unmeasurable. Mathematics 6, 119. doi: 10.3390/math6070119
Denger K., Cook A. M. (2010). Racemase Activity Effected by Two Dehydrogenases in Sulfolactate Degradation by Chromohalobacter Salexigens: Purification of (S)-Sulfolactate Dehydrogenase. Microbiol. (Read. Engl.) 156 (Pt 3), 967–974. doi: 10.1099/mic.0.034736-0
Devadatha B., Sarma V. V., Ariyawansa H., Jones E. G. (2018). Deniquelata Vittalii Sp. Nov., a Novel Indian Saprobic Marine Fungus on Suaeda Monoica and Two New Records of Marine Fungi From Muthupet Mangroves, East Coast of India. Mycosphere 9, 565–582. doi: 10.5943/mycosphere/9/3/8
Dube J. P., Valverde A., Steyn J. M., Cowan D. A., Van der Waals J. E. (2019). Differences in Bacterial Diversity, Composition and Function Due to Long-Term Agriculture in Soils in the Eastern Free State of South Africa. Diversity 11, 61. doi: 10.3390/d11040061
Empadinhas N., da Costa M. S. (2006). Diversity and Biosynthesis of Compatible Solutes in Hyper/Thermophiles. Int. Microbiol. 9 (3), 199–206. doi: 10.2436/20.7010.01.65
Ferry J. G. (2011). Fundamentals of Methanogenic Pathways that Are Key to the Biomethanation of Cbiomass. Curr. Opin. Biotechnol. 22 (3), 351–357.
Frank S., Brindley A. A., Deery E., Heathcote P., Lawrence A. D., Leech H. K., et al. (2005). Anaerobic Synthesis of Vitamin B12: Characterization of the Early Steps in the Pathway. Biochem. Soc. Trans. 33 (Pt 4), 811–814. doi: 10.1042/BST0330811
Friedmann S., Alber B. E., Fuchs G. (2006). Properties of Succinyl-Coenzyme A:D-Citramalate Coenzyme A Transferase and Its Role in the Autotrophic 3-Hydroxypropionate Cycle of Chloroflexus Aurantiacus. J. Bacteriol. 188 (18), 6460–6468. doi: 10.1128/JB.00659-06
Furtak K., Grządziel J., Gałązka A., Niedźwiecki J. (2019). Analysis of Soil Properties, Bacterial Community Composition, and Metabolic Diversity in Fluvisols of a Floodplain Area. Sustainability 11, 3929. doi: 10.3390/su11143929
García-López R., Cornejo-Granados F., Lopez-Zavala A. A., Sánchez-López F., Cota-Huízar A., Sotelo-Mundo R. R., et al. (2020). Doing More With Less: A Comparison of 16S Hypervariable Regions in Search of Defining the Shrimp Microbiota. Microorganisms 8, 134. doi: 10.3390/microorganisms8010134
Gardner D., Ralphs M., Turner D., Welsh S. (2002). Taxonomic Implications of Diterpene Alkaloids in Three Toxic Tall Larkspur Species (Delphinium Spp.). Biochem. Syst. Ecol. 30, 77–90. doi: 10.1016/S0305-1978(01)00120-X
Goswami D., Pithwa S., Dhandhukia P., Thakker J. N. (2014). Delineating Kocuria Turfanensis 2M4 as a Credible PGPR: A Novel IAA-Producing Bacteria Isolated From Saline Desert. J. Plant Interact. 9, 566–576. doi: 10.1080/17429145.2013.871650
Gunawardana D. (2020). An in Silico Study of Two Transcription Factors Controlling Diazotrophic Fates of the Azolla Major Cyanobiont Trichormus Azollae. Bioinf. Biol. Insights 14, 1177932220977490. doi: 10.1177/1177932220977490
Haase I., Sarge S., Illarionov B., Laudert D., Hohmann H.-P., Bacher A., et al. (2013). Enzymes From the Haloacid Dehalogenase (HAD) Superfamily Catalyse the Elusive Dephosphorylation Step of Riboflavin Biosynthesis. Chembiochem 14 (17), 2272–2275. doi: 10.1002/cbic.201300544
Harwood C. S., Burchhardt G., Herrmann H., Fuchs G.. (1998). Anaerobic Metabolism of Aromatic Compounds via the Benzoyl-CoA Pathway. FEMS Microbiol. Rev. 22 (5), 439–458.
Jeong J., Yun K., Mun S., Chung W. H., Choi S. Y., Nam Y. D., et al. (2021). The Effect of Taxonomic Classification by Full-Length 16S rRNA Sequencing With a Synthetic Long-Read Technology. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-90067-z
Jha Y., Subramanian R. B. (2018). “Effect of Root-Associated Bacteria on Soluble Sugar Metabolism in Plant Under Environmental Stress,” in Plant Metabolites and Regulation Under Environmental Stress (UK:Academic Press), 231–240.
Jiang J., Tetzlaff C. N., Takamatsu S., Iwatsuki M., Komatsu M., Ikeda H., et al. (2009). Genome Mining in Streptomyces Avermitilis: A Biochemical Baeyer-Villiger Reaction and Discovery of a New Branch of the Pentalenolactone Family Tree. Biochemistry 48 (27), 6431–6440. doi: 10.1021/bi900766w
Johnson C. D., Chary S. N., Chernoff E. A., Zeng Q., Running M. P., Crowell D. N. (2005). Protein Geranylgeranyltransferase I Is Involved in Specific Aspects of Abscisic Acid and Auxin Signaling in Arabidopsis. Plant Physiol. 139 (2), 722–733. doi: 10.1104/pp.105.065045
Joshi A., Kanthaliya B., Rajput V., Minkina T., Arora J. (2020). Assessment of Phytoremediation Capacity of Three Halophytes: Suaeda Monoica, Tamarix Indica and Cressa Critica. Biol. Futura 71, 301–312. doi: 10.1007/s42977-020-00038-0
Joulak I., Concórdio-Reis P., Torres C. A., Sevrin C., Grandfils C., Attia H., et al. (2021). Sustainable Use of Agro-Industrial Wastes as Potential Feedstocks for Exopolysaccharide Production by Selected Halomonas Strains. Environ. Sci. Poll Res. 29 (15), 22043–22055. doi: 10.1007/s11356-021-17207-w.
Karatan E., Duncan T. R., Watnick P. I. (2005). NspS, a Predicted Polyamine Sensor, Mediates Activation of Vibrio Cholerae Biofilm Formation by Norspermidine. J. Bacteriol. 187 (21), 7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005
Kawaguchi K., Senoura T., Ito S., Taira T., Ito H., Wasaki J., et al. (2014). The Mannobiose-Forming Exo-Mannanase Involved in a New Mannan Catabolic Pathway in Bacteroides Fragilis. Arch. Microbiol. 196 (1), 17–23. doi: 10.1007/s00203-013-0938-y
Kearl J., McNary C., Lowman J. S., Mei C., Aanderud Z. T., Smith S. T, et al. (2019). Salt-Tolerant Halophyte Rhizosphere Bacteria Stimulate Growth of Alfalfa in Salty Soil. Front. Microbiol. 10, 1849. doi: 10.3389/fmicb.2019.01849
Kim S., Lee S. B. (2006). Characterization of Sulfolobus Solfataricus 2-Keto-3-Deoxy-D-Gluconate Kinase in the Modified Entner-Doudoroff Pathway. Biosci. Biotechnol. Biochem. 70 (6), 1308–1316. doi: 10.1271/bbb.50566
Klatt C. G., Bryant D. A., Ward D. M. (2007). Comparative Genomics Provides Evidence for the 3-Hydroxypropionate Autotrophic Pathway in Filamentous Anoxygenic Phototrophic Bacteria and in Hot Spring Microbial Mats. Environ. Microbiol. 9 (8), 2067–2078. doi: 10.1111/j.1462-2920.2007.01323.x
Köberl M., Schmidt R., Ramadan E. M., Bauer R., Berg G. (2013). The Microbiome of Medicinal Plants: Diversity and Importance for Plant Growth, Quality and Health. Front. Microbiol. 4, 400. doi: 10.3389/fmicb.2013.00400
Koga Y., Nakano M. (2008). A Dendrogram of Archaea Based on Lipid Component Parts Composition and Its Relationship to rRNA Phylogeny. Systematic Appl. Microbiol. 31 (3), 169–182. doi: 10.1016/j.syapm.2008.02.005
Kurth J. M., Op denCamp H. J., Welte C. U (2020). Several Ways One Goal—Methanogenesis From Unconventional Substrate. Appl. Microbiol. Biotechnol. 104 (16), 6839–6854.
Kuznetsova A.I., Ivanova E. A., Samylina O. S., Kurbanova F. G., Gruzdev D. S., Kanapatskiy T. A, et al. (2020). Prokaryotic Communities in Saline Soils of the Lake Elton Area in a Soil Catena Along the Khara River. Microbiology 89, 670–684. doi: 10.1134/S0026261720060119
Lacap D. C., Warren-Rhodes K. A., McKay C. P., Pointing S. B. (2011). Cyanobacteria and Chloroflexi-Dominated Hypolithic Colonization of Quartz at the Hyper-Arid Core of the Atacama Desert, Chile. Extremophiles 15, 31–38. doi: 10.1007/s00792-010-0334-3
Lena T., Suda W. (2018). “Mapping the Environmental Microbiome,” in Encyclopedia of Bioinformatics and Computational Biology. Eds. R S., G M., N K., S C. (Netherlands: Elsevier B.V.), pp. 17–28.
Lin S.-J., Ford E., Haigis M., Liszt G., Guarente L. (2004). Calorie Restriction Extends Yeast Life Span by Lowering the Level of NADH. Genes Dev. 18 (1), 12–16. doi: 10.1101/gad.1164804
Liu H., Brettell L. E. (2019). Plant Defense by VOC-Induced Microbial Priming. Trends Plant Sci. 24, 187–189. doi: 10.1016/j.tplants.2019.01.008
Löffler M., Fairbanks L. D., Zameitat E., Marinaki A. M., Simmonds H. A. (2005). Pyrimidine Pathways in Health and Disease. Trends Mol. Med. 11 (9), 430–437. doi: 10.1016/j.molmed.2005.07.003
Looman J. (1984). Biological Flora of Canada. 5. Delphinium Glaucum Watson, Tall Larkspur. Can. Field-Nat. 98 (3), 345–361.
MacDonald Z. G., Nielsen S. E., Acorn J. H. (2017). Negative Relationships Between Species Richness and Evenness Render Common Diversity Indices Inadequate for Assessing Long-Term Trends in Butterfly Diversity. Biodiversity Conserv. 26, 617–629. doi: 10.1007/s10531-016-1261-0
Maeng S., Park Y., Lee S. E., Han J. H., Cha I. T., Lee K. E, et al. (2020). Pontibacter Pudoricolor Sp. Nov., and Pontibacter Russatus Sp. Nov. Radiation-Resistant Bacteria Isolated From Soil. Antonie Van Leeuwenhoek 113, 1361–1369. doi: 10.1007/s10482-020-01446-x
Maheshwari D. K., Saraf M. (2016). Halophiles: Biodiversity and Sustainable Exploitation (Sustainable Development and Biodiversity, 6) (Softcover Reprint of the Original. 1st ed. 2015 ed Vol. 6 (Switzerland:Springer).
Malaviya A., Gomes J. (2008). Androstenedione Production by Biotransformation of Phytosterols. Bioresource Technol. 99 (15), 6725–6737. doi: 10.1016/j.biortech.2008.01.039
Manandhar M., Cronan J. E. (2017). Pimelic Acid, the First Precursor of the Bacillus Subtilis Biotin Synthesis Pathway, Exists as the Free Acid and Is Assembled by Fatty Acid Synthesis. Mol. Microbiol. 104 (4), 595–607. doi: 10.1111/mmi.13648
Manners G. D., Panter K. E., Pfister J. A., Ralphs M. H., James L. F. (1998). The Characterization and Structure− Activity Evaluation of Toxic Norditerpenoid Alkaloids From Two Delphinium Species. J. Nat. Prod. 61, 1086–1089. doi: 10.1021/np980013e
McGovern E., Waters S. M., Blackshields G., McCabe M. S. (2018). Evaluating Established Methods for Rumen 16S rRNA Amplicon Sequencing With Mock Microbial Populations. Front. Microbiol. 9, 1365. doi: 10.3389/fmicb.2018.01365
Mehnaz D., Mukhtar S., Ishaq A., Hassan S., Abdulla K., Mirza M. S. (2017). Comparison of Microbial Communities Associated With Halophyte (Salsola Stocksii) and Non-Halophyte (Triticum Aestivum) Using Culture-Independent Approaches. Pol. J. Microbiol. 66 (3), 375–386. doi: 10.5604/01.3001.0010.4866
Morris E. K., Caruso T., Buscot F., Fischer M., Hancock C., Maier T. S, et al. (2014). Choosing and Using Diversity Indices: Insights for Ecological Applications From the German Biodiversity Exploratories. Ecol. Evol. 4, 3514–3524. doi: 10.1002/ece3.1155
Muthazhagan K., Thirunavukkarasu P., Ramanathan T., Kannan D. (2014). Studies on Phytochemical Screening, Antimicrobial and Anti Radical Scavenging Effect Coastal Salt Mash Plant of a Suaeda Monoica. Res. J. Phytochem. 8, 102–111. doi: 10.3923/rjphyto.2014.102.111
Nunn C. E. M., Johnsen U., Schönheit P., Fuhrer T., Sauer U., Hough D. W., et al. (2010). Metabolism of Pentose Sugars in the Hyperthermophilic Archaea Sulfolobus Solfataricus and Sulfolobus Acidocaldarius. J. Biol. Chem. 285 (44), 33701–33709. doi: 10.1074/jbc.M110.146332
Rabus R., Boll M., Heider J., Meckenstock R. U., Buckel W., Einsle O., et al. (2016). Anaerobic Microbial Degradation of Hydrocarbons: From Enzymatic Reactions to the Environment. Microb. Physiol. 26 (1–3), 5–28.
Rajathi F. A. A., Arumugam R., Saravanan S., Anantharaman P. (2014). Phytofabrication of Gold Nanoparticles Assisted by Leaves of Suaeda Monoica and Its Free Radical Scavenging Property. J. Photochem. Photobiol. B.: Biol. 135, 75–80. doi: 10.1016/j.jphotobiol.2014.03.016
Rast P., Glöckner I., Boedeker C., Jeske O., Wiegand S., Reinhardt R., et al. (2017). Three Novel Species With Peptidoglycan Cell Walls Form the New Genus Lacunisphaera Gen. Nov. In the Family Opitutaceae of the Verrucomicrobial Subdivision 4. Front. Microbiol. 8, 202. doi: 10.3389/fmicb.2017.00202
Rochman F. F., Kim J. J., Rijpstra W. I.C., Damsté J. S. S., Schumann P., Verbeke T. J., et al. (2018). Oleiharenicola Alkalitolerans Gen. Nov., Sp. Nov., a New Member of the Phylum Verrucomicrobia Isolated From an Oilsands Tailings Pond. Int. J. Syst. Evol. Microbiol. 68, 1078–1084. doi: 10.1099/ijsem.0.002624
Selesi D., Meckenstock R. U.. (2009). Anaerobic Degradation of the Aromatic Hydrocarbon Biphenyl by a Sulfate-Reducing Enrichment Culture. FEMS Microbiol. Ecol. 68 (1), 86–93.
Shaheen U., Shoeib N. A., Temraz A., Abdelhady M. I. (2017). Flavonoidal constituents, antioxidant, antimicrobial, and cytotoxic activities of dipterygium glaucum grown in kingdom of Saudi Arabia. Pharmacogn. Mag. 13 (Suppl 3), S484. doi: 10.4103/pm.pm_44_16
Shaw F. L., Elliott K. A., Kinch L. N., Fuell C., Phillips M. A., Michael A. J. (2010). Evolution and Multifarious Horizontal Transfer of an Alternative Biosynthetic Pathway for the Alternative Polyamine Sym-Homospermidine. J. Biol. Chem. 285 (19), 14711–14723. doi: 10.1074/jbc.M110.107219
Shoji M., Sato K., Yukitake H., Naito M., Nakayama K. (2014). Involvement of the Wbp Pathway in the Biosynthesis of Porphyromonas Gingivalis Lipopolysaccharide With Anionic Polysaccharide. Sci. Rep. 4, 5056. doi: 10.1038/srep05056
Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., et al. (2010). Bacterial Phenylalanine and Phenylacetate Catabolic Pathway Revealed. Proc. Natl. Acad. Sci. U. S. A. 107 (32), 14390–14395. doi: 10.1073/pnas.1005399107
Thweatt J. L., Canniffe D. P., Bryant D. A. (2019). Biosynthesis of Chlorophylls and Bacteriochlorophylls in Green Bacteria. Adv. Bot. Res. 90, 35–89. doi: 10.1016/bs.abr.2019.03.002
Umitsu M., Nishimasu H., Noma A., Suzuki T., Ishitani R., Nureki O. (2009). Structural Basis of AdoMet-Dependent Aminocarboxypropyl Transfer Reaction Catalyzed by tRNA-Wybutosine Synthesizing Enzyme, TYW2. Proc. Natl. Acad. Sci. U. S. A. 106 (37), 15616–15621. doi: 10.1073/pnas.0905270106
Waite D. W., Chuvochina M., Pelikan C., Parks D. H., Yilmaz P., Wagner M., et al. (2020). Proposal to Reclassify the Proteobacterial Classes Deltaproteobacteria and Oligoflexia, and the Phylum Thermodesulfobacteria Into Four Phyla Reflecting Major Functional Capabilities. Int. J. Syst. Evol. Microbiol. 70, 5972–6016. doi: 10.1099/ijsem.0.004213
Wang Y. X., Liu J. H., Xiao W., Ma X. L., Lai Y. H., Li Z. Y, et al. (2013). Aliifodinibius Roseus Gen. Nov., Sp. Nov., and Aliifodinibius Sediminis Sp. Nov., Two Moderately Halophilic Bacteria Isolated From Salt Mine Samples. Int. J. Syst. Evol. Microbiol. 63, 2907–2913. doi: 10.1099/ijs.0.043869-0
Wang W., Shi L., Qin Y., Li F.. (2020). Research and Application of Chondroitin Sulfate/dermatan Sulfate-Degrading Enzymes. Front. Cell Dev. Biol. 1435.
Ward L. M., Hemp J., Shih P. M., McGlynn S. E., Fischer W. W. (2018). Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer. Front. Microbiol. 9, 260. doi: 10.3389/fmicb.2018.00260
Watanabe S., Piyanart S., Makino K. (2008). Metabolic Fate of L-Lactaldehyde Derived From an Alternative L-Rhamnose Pathway. FEBS J. 275 (20), 5139–5149. doi: 10.1111/j.1742-4658.2008.06645.x
Welsh S., Ralphs M. (2002). Some Tall Larkspurs (Delphinium-Ranunculaceae) a Taxonomic Review. Biochem. Syst. Ecol. 2, 103–112. doi: 10.1016/S0305-1978(01)00122-3
Willis C., Desai D., LaRoche J. (2019). Influence of 16S rRNA Variable Region on Perceived Diversity of Marine Microbial Communities of the Northern North Atlantic. FEMS Microbiol. Lett. 366, fnz152. doi: 10.1093/femsle/fnz152
Wolińska A. (2019). Metagenomic Achievements in Microbial Diversity Determination in Croplands: A Review. Microb. Diversity Genom. Era, 15–35. doi: 10.1016/B978-0-12-814849-5.00002-2
Wu X., Li J., Ji M., Wu Q., Wu X., Ma Y., et al. (2019). Non-Synchronous Structural and Functional Dynamics During the Coalescence of Two Distinct Soil Bacterial Communities. Front. Microbiol. 10, 1125. doi: 10.3389/fmicb.2019.01125
Yokooji Y., Tomita H., Atomi H., Imanaka T. (2009). Pantoate Kinase and Phosphopantothenate Synthetase, Two Novel Enzymes Necessary for CoA Biosynthesis in the Archaea. J. Biol. Chem. 284 (41), 28137–28145. doi: 10.1074/jbc.M109.009696
Yurgel S. N., Nearing J. T., Douglas G. M., Langille M. G. (2019). Metagenomic Functional Shifts to Plant Induced Environmental Changes. Front. Microbiol. 10, 1682. doi: 10.3389/fmicb.2019.01682
Zhao X., Miao S., Sun Y., Gong Q., Zhao J., Wang J, et al. (2020). Aliifodinibius Salipaludis Sp. Nov., Isolated From Saline-Alkaline Soil. Curr. Microbiol. 77, 1328–1333. doi: 10.1007/s00284-019-01863-w
Keywords: soil, microbiome, V3–V4, Suaeda monoica, Dipterygium glaucum
Citation: Jalal RS, Sheikh HI, Alotaibi MT, Shami AY, Ashy RA, Baeshen NN, Abulfaraj AA, Baz L, Refai M, Baeshen NA, Fadhlina A, Arifullah M and Baeshen MN (2022) The Microbiome of Suaeda monoica and Dipterygium glaucum From Southern Corniche (Saudi Arabia) Reveals Different Recruitment Patterns of Bacteria and Archaea. Front. Mar. Sci. 9:865834. doi: 10.3389/fmars.2022.865834
Received: 30 January 2022; Accepted: 29 March 2022;
Published: 28 April 2022.
Edited by:
Siddhartha Pati, NatNov Bioscience Pvt Ltd., IndiaReviewed by:
Nilesh Prakash Nirmal, Mahidol University, ThailandManzoor A. Mir, University of Kashmir, India
Copyright © 2022 Jalal, Sheikh, Alotaibi, Shami, Ashy, Baeshen, Abulfaraj, Baz, Refai, Baeshen, Fadhlina, Arifullah and Baeshen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hassan I. Sheikh, sheikhinho@gmail.com
 Rewaa S. Jalal1
Rewaa S. Jalal1  Hassan I. Sheikh
Hassan I. Sheikh Ashwag Y. Shami
Ashwag Y. Shami Naseebh N. Baeshen
Naseebh N. Baeshen Aala A. Abulfaraj
Aala A. Abulfaraj Mohammed Refai
Mohammed Refai Mohammed N. Baeshen
Mohammed N. Baeshen