Net diversification rates of the woody plant genus Petalidium (Acanthaceae) are highest in the ancient and arid Namib Desert
- 1Crew Building, School of Geosciences, University of Edinburgh, Edinburgh, United Kingdom
- 2Department of Ecology and Evolutionary Biology, University of Colorado, Boulder, CO, United States
- 3Independent Researcher, Windhoek, Namibia
- 4Royal Botanic Garden Edinburgh, Edinburgh, United Kingdom
At present, tropical arid biomes house less woody plant species diversity than tropical moist biomes, which could be due to lower rates of evolutionary diversification in the recent or distant past. Here, we study the evolutionary diversification of Petalidium (Acanthaceae), a genus of 36 species of woody shrubs found in the Namib Desert of southwest Africa, and surrounding areas. We generated a new, nearly fully sampled and temporally calibrated phylogeny for Petalidium using RADseq SNP data and secondary calibrations. We then investigated variation in net diversification rate across the phylogeny, the ancestral climatic niche of lineages and the link between the two. We find that arid climatic conditions are linked with increased rates of net species diversification in the genus. Despite its great age, the Namib Desert clearly hosts young plant radiations. This apparent contradiction can be explained by a scenario of high evolutionary turnover, in this case potentially caused by alternating hyper-arid and relatively mesic phases. Hyper-arid phases could result in high plant mortality and extinction of species, leading to ecological opportunity and diversification during mesic phases. Taken together, our results contribute to a growing body of literature that shows evidence for elevated rates of plant diversification in the Quaternary in arid biomes across the globe.
Introduction
Within the tropics, the diversity of plant species is generally linked to water availability, with more moist biomes housing higher taxonomic diversity, particularly for woody plants (O'Brien et al., 2000; Daru et al., 2016; Esquivel-Muelbert et al., 2017). Within individual woody plant clades this pattern is also evident, with families and genera tending to be more species-rich in moist biomes compared to more arid biomes (Currie, 1991; Pennington et al., 2006). Assuming no systematic difference in the age of clades, these discrepancies could suggest that diversification rates are lower in arid versus moist biomes.
In contrast, scientists since Stebbins (1952) have argued that arid environments promote plant speciation (Axelrod, 1972; Böhnert et al., 2022). Arid biomes may house a greater diversity of edaphic conditions that can facilitate plant speciation (Stebbins, 1952; Axelrod, 1972; Buira et al., 2021), and the patchy distribution of species in arid biomes may be conducive to intermediate levels of gene flow among populations that can promote adaptive diversity and differentiation (Wright, 1940; Stebbins, 1952).
Present-day arid biomes have a dynamic climatological past, and increasing aridity since the Miocene across the globe has been cited as a causal factor in diversification of arid-affiliated plant lineages (Wu et al., 2018; Böhnert et al., 2022; Čalasan et al., 2022; Hua et al., 2022). Global aridification continued to increase throughout the Pliocene and Pleistocene climate cycles, with multiple examples of Quaternary diversification in arid-affiliated plant lineages (Kadereit and Abbott, 2021). There is ample evidence that arid biomes are areas of active and recent plant speciation. However, most studied examples pertain to herbaceous and succulent plant lineages (e.g., Klak et al., 2004; Arakaki et al., 2011; Guerrero et al., 2013; Böhnert et al., 2019; Lavor et al., 2019; Merklinger et al., 2021). Macroevolutionary studies of woody plant lineages, which show the most marked differences in diversity between arid and moist biomes, are scarce (although see Simon et al., 2009; Singhal et al., 2021). We do not yet have a firm idea of the relationship between arid climatic conditions and diversification in woody plant lineages.
Sub-Saharan Africa as a whole is dry in comparison to other regions of the tropics, such as the Americas and southeast Asia. Yet, arid regions of sub-Saharan Africa have received limited macroevolutionary study compared to regions like the Atacama Desert (Guerrero et al., 2013; Böhnert et al., 2019; Merklinger et al., 2021). The Namib Desert, in southwest Africa, is one of the oldest deserts on Earth, originating 55–80 Ma (Ward et al., 1983). It houses ancient iconic plant lineages such as the morphologically bizarre gymnosperm Welwitschia (Welwitschiaceae) and the similarly bizarre angiosperm lineage Tiganophyton (Tiganophytaceae, Swanepoel et al., 2020). These species-poor lineages are notable for occurring on long, isolated evolutionary branches, which have been termed “depauperons” (Donoghue and Sanderson, 2015). In contrast, other lineages emblematic of the Namib Desert have achieved markedly high taxonomic diversity, such as Euphorbia (Euphorbiaceae), Indigofera (Fabaceae), Commiphora (Burseraceae) and several genera of Acanthaceae (Craven, 2009; Darbyshire et al., 2020). Most of these clades are diverse across tropical Africa, and the species present in the hyper-arid region of southwest Africa are derived from multiple lineages.
One genus of Acanthaceae, Petalidium, stands out from other diverse genera in the region in that its diversity is centred in the Namib Desert and surrounding areas, and is hypothesized to derive from a single colonisation event (Tripp et al., 2017). Petalidium is furthermore a lineage of woody, non-succulent plants, a habit usually associated with higher diversity in moist regions. In the present study, we aim to test whether the arid conditions in the Namib Desert may have helped to spur diversification in the genus Petalidium, as proposed by Tripp et al. (2017). If supported, Petalidium would represent a notable counter-example to the assumption that tropical arid biomes inhibit woody plant diversification.
Petalidium is a genus of 36 accepted species that occur almost entirely in Namibia and Angola, with the exception of one species sister to the African taxa, which occurs in India and Nepal in a more moist climate and two species restricted to northeastern South Africa and adjacent areas in Zimbabwe (Tripp et al., 2017). Petalidium is the fifth most diverse genus in the arid Kaokoveld Region of northwestern Namibia and southwestern Angola (Craven, 2009) and is ecologically successful, being the dominant shrub cover in major portions of this region and also forming large populations of co-occurring species in many areas (all authors, pers. obs.). The total taxonomic diversity of Petalidium is likely higher than currently known as new species of Petalidium continue to be found after fieldwork in poorly studied regions (Swanepoel, 2020; Swanepoel and Manzitto-Tripp, 2022). African Petalidium appear to be undergoing an evolutionary radiation, with estimates for the crown age of the 35 African species ranging from 1.4 Ma to 3.6 Ma (Tripp et al., 2017). Species of Petalidium can occur in extremely arid areas where mean annual precipitation (MAP) is less than 50 mm per year, but other species of African Petalidium occur in more moist areas where MAP exceeds 700 mm. We know that at present the majority of species of Petalidium occur in more arid regions (Tripp et al., 2017), but the climatic history of the clade has not been studied. We do not know whether it originated in more moist or arid conditions. If it originated in more arid conditions, then its greater diversity in arid regions may simply be a result of a longer history of occupancy.
In order to examine the link between climatic conditions and diversification in Petalidium, we undertook the following steps: (i) we generated a new phylogeny for the genus using RAD sequencing, improving the species sampling from previous efforts by 50% (Tripp et al., 2017); (ii) we assessed variation in net diversification rate across the phylogeny; (iii) we reconstructed ancestral climatic niches across the phylogeny; and (iv) we tested for a link between net diversification and estimated ancestral climatic niche. Our overall aim was to test whether Petalidium shows higher species richness in arid climatic conditions relative to moist ones because of higher rates of net diversification in arid conditions or because of a longer history of occupancy of arid climatic environments, with equivalent or even lower net diversification in arid conditions.
Materials and methods
Ecological analyses
We compiled 1,349 occurrence records of Petalidium from herbarium collections. We manually checked this database, verified specimens’ identifications and removed those that could not be confidently identified or geo-referenced. We then extracted values for the 19 Worldclim bioclimatic variables (Fick and Hijmans, 2017) for remaining occurrences. We computed species means for all variables and performed a principal component analysis (PCA) on mean environmental preference using the princomp function (on the correlation matrix) in the base package of the R Statistical Environment (R Core Team, 2013).
Field methods
Populations of Petalidium were sampled for herbarium and silica-gel vouchers during four expeditions to Namibia, one to South Africa and one to Angola, by E.A.T and K.G.D. from 2010 to 2017. Specimens are housed at the following herbaria: RSA-POM, COLO, WIND, LUBA, K, and E. All specimens included in this phylogenetic work were fertile, and we are confident in the identifications.
DNA extraction and sequencing
For 35 samples, we extracted DNA from silica-dried leaf tissue using a standard CTAB protocol (Doyle and Doyle, 1987). DNA was then sequenced using a ddRADseq approach (see Tripp et al., 2017 for protocols including custom barcode design, which was modified from Parchman et al., 2012). We used EcoR1 and Mse1 for double digestion followed by size selection of 250–500 bp fragments to reduce the overall genomic material that was sequenced. Libraries were pooled and then submitted to the University of Colorado’s BioFrontiers Sequencing Facility for quality control, cleanup, and 1 × 100 sequencing on an Illumina NextSeq.
Bioinformatics
All bioinformatic work was conducted on the University of Colorado’s SUMMIT supercomputer, which comprised a total of 450 compute nodes, each containing 24 cores, for a total of 10,800 available cores (each with 200 GB of RAM). We used FastQC (Andrews, 2017) to assess overall quality of our sequencing. Subsequently, the data were demultiplexed using ea. fastq-multx v.1.03, which is distributed as part of the ea-utils package (Aronesty, 2011). Then, adapters were removed and reads were cleaned using cutadapt v.1.4.2 (Martin, 2011). The resulting fastq files were assessed for total quantity of reads containing the CAATT cutsite in Unix, and any samples with fewer than 100,000 reads with the cutsite were omitted from further consideration. We used ipyrad (Eaton and Overcast, 2020) to trim low-quality bases, filter reads and then assemble the data into loci by implementing reference-guided methods, leveraging a fully assembled, although not yet published nuclear genome for Petalidium crispum (Manzitto-Tripp et al., unpub. data). The following parameters were implemented in ipyrad: max_low_qual_bases = 10, mindepth_majrule = 2; cust_threshold = 0.85; max_Ns_consens = 5; max_Hs_consens = 25; min_samples_locus = 25; max_Indels_locus = 100; and max_shared_Hs_locus = 0.5. Paralogous loci were excluded from consideration using the max_shared_Hs_locus = 2 flag. The final RADseq phylogenomic dataset is available on GenBank under the Sequence Read Archive Study SUB12441993.
Phylogenetic reconstruction and divergence time estimation
We used the matrix of SNPs to infer phylogenetic relationships under the GTR model of substitution using maximum likelihood (ML) in RaxML (Stamatakis, 2014) with 100 bootstrap replicates. We included SNPs that were missing data for up to 90% of the accessions in the analysis. We explored lower thresholds of missing data, but found that the 90% level gave the strongest bootstrap support values (as in Tripp et al., 2017). To avoid bias in branch length estimation due to the absence of constant sites, we applied the ascertainment bias correction to likelihood calculations (Leaché et al., 2015). We also ran a phylogenetic analysis in IQ-TREE multicore version 2.0.3 (Nguyen et al., 2015) under the GTR model of substitution and the ascertainment bias correction, with 1,000 ultrafast bootstrap replicates (Hoang et al., 2018). Missing data in our matrix of SNPs precluded the use of SNAPP, a Bayesian method implemented in BEAST2 that can simultaneously infer tree topology and node ages (Bouckaert et al., 2014). Instead, we dated the ML phylogenetic tree to absolute time using penalized-likelihood, implemented in treePL (Smith and O’Meara, 2012). We set a secondary calibration using an interval of 1.3–5 Ma corresponding to the 95%HPD of the crown age of Petalidium estimated by Tripp et al. (2017). We used cross-validation to estimate the best value of the smoothing parameter. There were two species (P. ohopohense and P. lanatum) for which sequence data were available from earlier RADseq runs, but with insufficient sequence data to place in the phylogeny in our complete phylogenetic analysis. Preliminary analyses with different sets of data allowed us to develop a robust idea of whom their closest relative(s) were. These two species were grafted into the final phylogeny mid-way along the branch subtending their closest relative(s) in R using the function bind.tip from the package phytools 1.2.0 (Revell, 2012).
Estimation of ancestral character states and diversification rate
We mapped the logarithm of precipitation of driest quarter along the phylogeny and conducted ancestral state estimation using the function contmap of the package phytools v1.2.0 (Revell, 2012), which in turn relies on the approach of Felsenstein (1985). Of the readily available bioclimatic variables, this one best mechanistically quantifies water availability to plants when it is most scarce, across the driest 3 months of the year. Precipitation of the driest month may be influenced by lags in depletion of water from preceding months, while precipitation in wetter periods may not accurately measure stress from low water availability.
We estimated the net diversification rate of Petalidium in a bayesian framework using the software BAMM v.2.5.0. (Rabosky, 2014). BAMM was run with a global sampling fraction of 35/36 and for 10 million generations. Convergence of the mcmc chains was checked in R using the coda package v0.19.4 (Plummer et al., 2008) and plots were made with the BAMMtools package v2.1.10 (Rabosky et al., 2014).
We tested for an association between aridity and net diversification rate using the Quantitative State Speciation and Extinction model (QuaSSE, FitzJohn, 2010), implemented in the R package diversitree v0.9.16 (FitzJohn, 2012). We ran QuaSSE on the first principal component of the climate space occupied by species, which broadly represents a gradient in water availability, as well as on the logarithm of precipitation of the driest quarter. We compared a null model of trait-independent diversification vs. models where the diversification rate has a linear, sigmoidal or hump-shaped response to the environmental variable, with and without drift included. Because trait-dependent diversification models have been shown to have an elevated type-I error rate (Rabosky and Goldberg, 2015), we conducted parametric bootstrapping to evaluate the significance of our results. First, we inferred the root state of the genus for the two variables using the function fastANC from the phytools package v1.2.0 (Revell, 2012). We then used the estimated root state and the estimate of the diffusion parameter from the null model in QuaSSE to perform 100 simulations of Brownian-motion trait evolution along the phylogeny, using the function sim.char from the geiger R package v2.0.10 (Harmon et al., 2007; Pennell et al., 2014). Finally, these 100 simulated trait histories were subjected to a QuaSSE model comparison as with the observed empirical data set. The log-likelihood of the best model minus the log likelihood of the null model (hereafter ΔLL) was recorded for each simulated dataset and compared to the observed ΔLL. The proportion of simulated ΔLL values that were greater than the observed ΔLL value was then calculated to assess the robustness of QuaSSE results. If less than 0.05 of the simulated values are greater than the observed value, then the observed value is considered to be significantly different from that expected by chance in this 1-tailed test.
Results
Ecological analyses
After cleaning the occurrence data, we obtained 982 occurrence records for 35 species. The first two principal components (PC1 and PC2) from a principal component analysis of the mean climatic variables for each species explained 48.1 and 27.8% of the variation in the climate data respectively, with subsequent axes explaining little variation (Figure 1). PC1 was related primarily to amounts of precipitation (total annual precipitation and total precipitation for different quarters and months), while PC2 was related primarily with mean temperature variables (annual, quarterly and monthly).
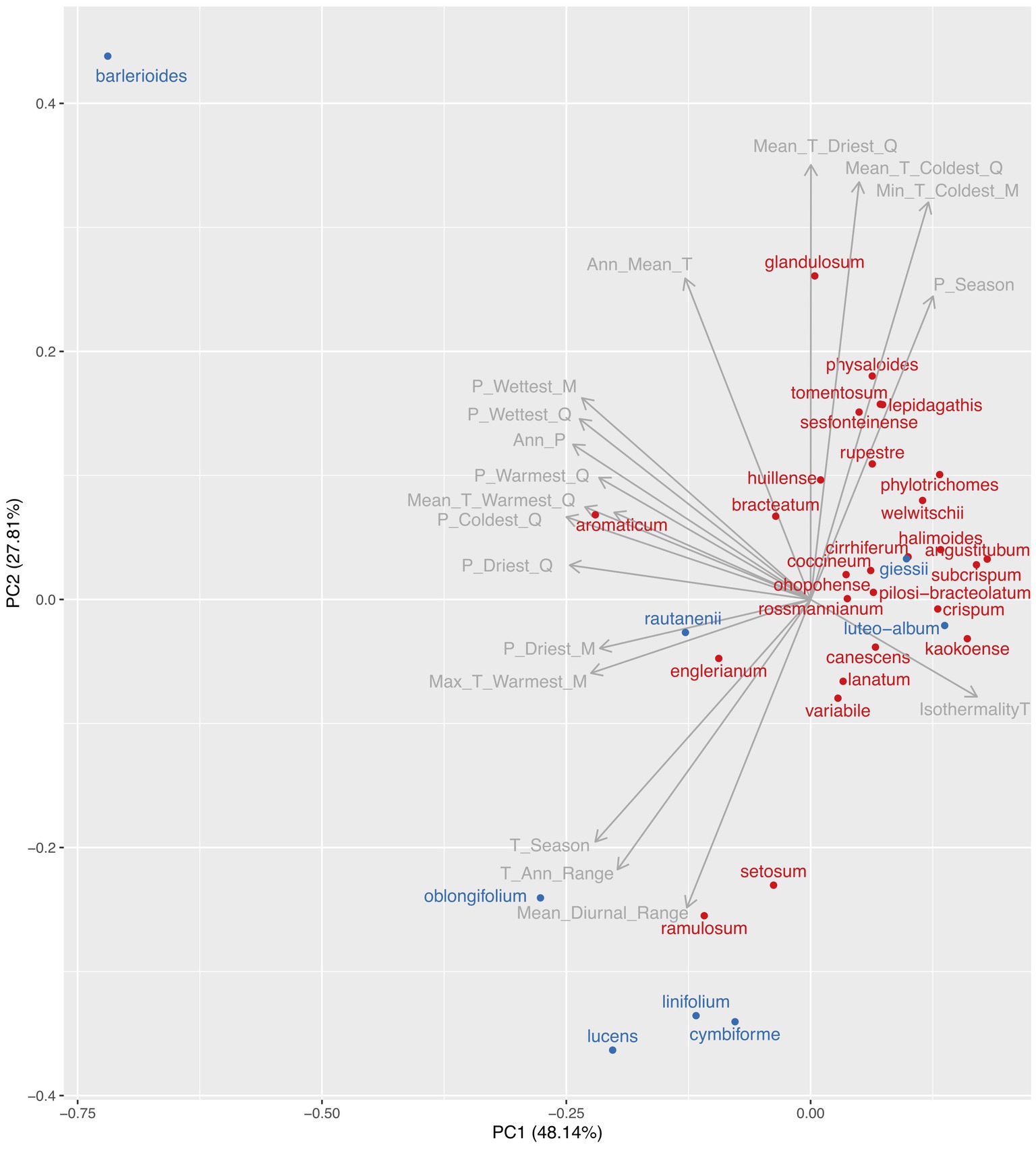
Figure 1. Position of species in a principal component analysis of the mean value for each species for 19 bioclimatic variables (from www.worldclim.org). The first two components are shown, which explain 48.1 and 27.8% of the variation in the data, respectively. Species from the “diverse and arid clade” which shows a faster rate of net diversification (see Figure 4) are denoted in red, and all other species are denoted in blue. The length of the arrow for each climatic variable is related to the strength of its correlation with the first two principal components. Climatic variables are abbreviated: Mean_Diurnal_Range = mean diurnal temperature range; T_Ann_Range = temperature annual range; T_Season = temperature seasonality; Max_T_Warmest_M = maximum temperature of the warmest month; P_Driest_M = total precipitation of the driest month; P_Driest_Q = total precipitation of the driest quarter; P_Coldest_Q = total precipitation of the coldest quarter; Mean_T_Warmest_Q = mean temperature of the warmest quarter; P_Warmest_Q = total precipitation of the warmest quarter; Ann_P = total annual precipitation; P_Wettest_Q = mean temperature of the wettest quarter; P_Wettest_M = mean temperature of the wettest month; Ann_Mean_T = mean annual temperature; Mean_T_Driest_Q = mean temperature of the driest quarter; Mean_T_Coldest_Q = mean temperature of the coldest quarter; Mean_T_Coldest_M = mean temperature of the coldest month; P_Season = precipitation seasonality; and IsothermalityT = isothermality. We interpret the first principal component as a temperature-dominated axis, with positive values being hotter and negative values being colder. We interpret the second principal component as a precipitation-dominated axis, with positive values being drier and negative values being wetter.
Sequencing and bioinformatics
After sequencing and bioinformatic processing, we obtained 83,547 SNPs. Prior to phylogenetic inference with an ascertainment bias correction, only true SNPs (i.e., sites with two bases present across all individuals) were retained, while all sites with one base or an IUPAC character, which are treated as invariants by many softwares including RaxML, were removed, resulting in a final matrix of 34,017 SNPs for 35 species.
Phylogenetics, ancestral state reconstruction and diversification analyses
RaxML (Figure 2) and IQ-TREE (Supplementary Figure S1) analyses yielded highly similar phylogenetic trees with similar support values. Deep nodes in the ML phylogenetic tree were fully resolved, but bootstrap support values were low within one clade. That clade holds the majority of Petalidium species, yet has the shortest branch lengths (Figure 2). Our temporally calibrated phylogeny had an estimated crown age for African Petalidium of 4.33 Ma. The clade with short branch lengths and most Petalidium species had an estimated crown age of 1.63 Ma (Figure 3).
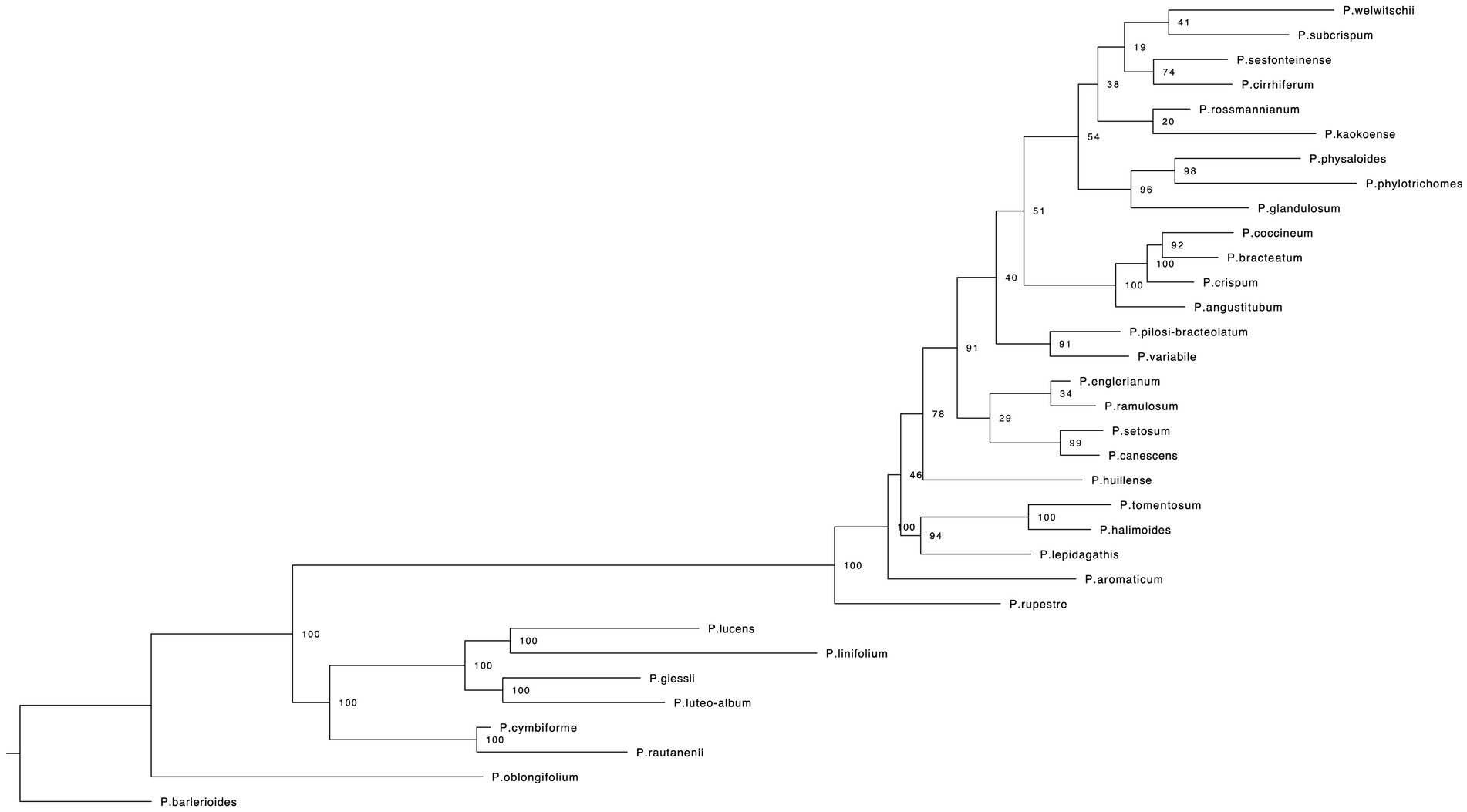
Figure 2. Maximum likelihood phylogeny for the genus Petalidium (Acanthaceae) from a RAxML analysis of RADseq genetic data (see Section Materials and methods for details). Numbers at nodes indicate support values from a maximum likelihood bootstrap analysis.
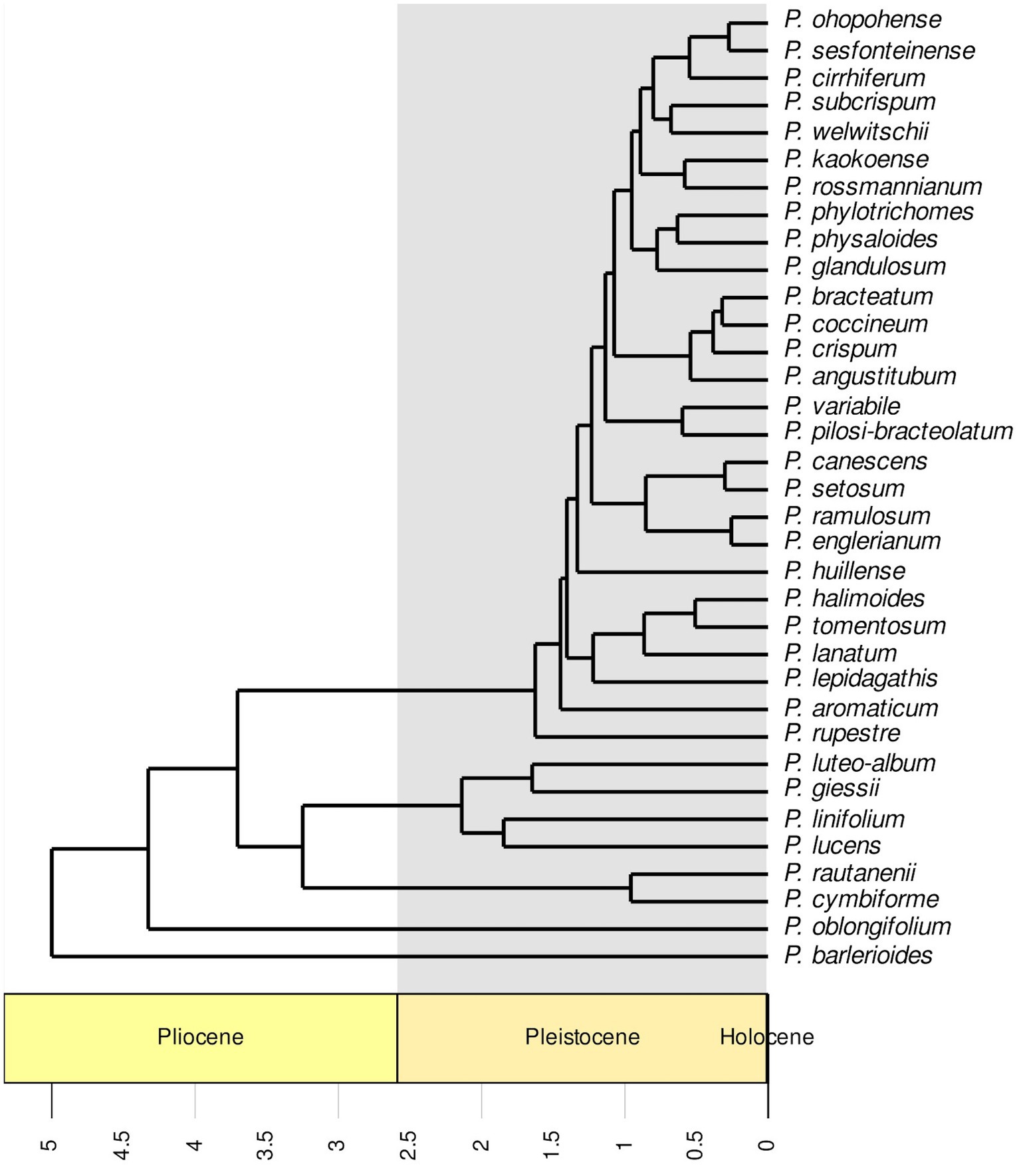
Figure 3. Temporally calibrated phylogeny of the genus Petalidium (Acanthaceae) generated from a maximum likelihood phylogeny (Figure 2) with a penalised likelihood analysis using secondary calibrations from a previous study Tripp et al. (2017). The timescale is in millions of years, with the Pleistocene shaded in grey. Most diversification events took place in the Pleistocene.
Ancestral state reconstruction of precipitation of the driest quarter suggests that Petalidium originated in more moist conditions and adapted to high levels of aridity in its recent evolutionary history (Figure 4). There are two lineages found within arid areas: one comprises two species (P. luteo-album and P. giessii). The other is the clade mentioned above with short branch lengths and comprising the majority of species in the genus. The first branch in this clade belongs to P. rupestre, a local endemic in the arid Namib Desert of southwest Angola, while the second branch belongs to P. aromaticum, a species occurring in more moist conditions along the Limpopo River on the border of South Africa and Zimbabwe. The remaining species in this clade occur in the arid Namib Desert and Kaokoveld. We hereafter refer to this clade as the diverse arid clade.
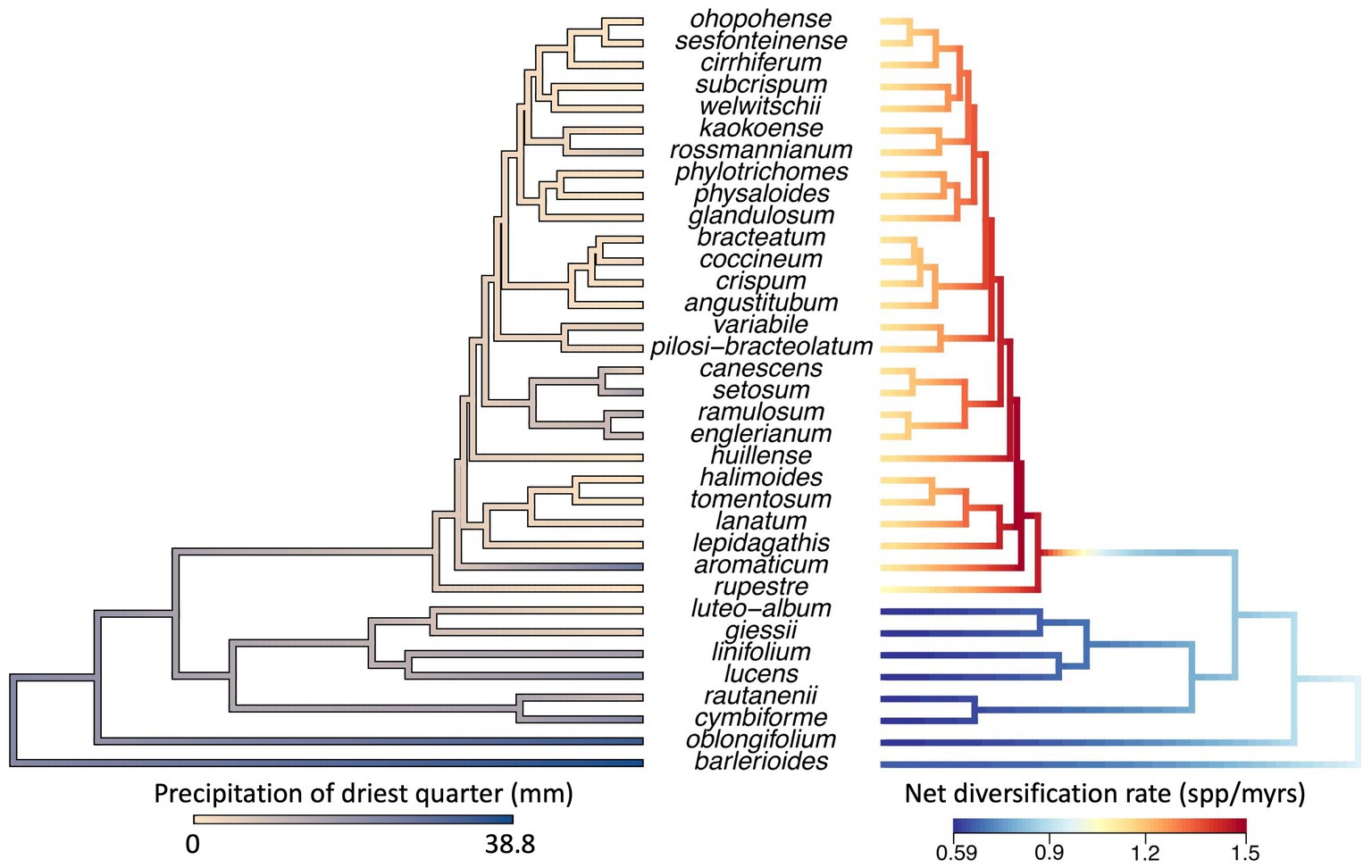
Figure 4. Left: Ancestral state reconstruction of precipitation of driest quarter for the genus Petalidium (Revell, 2012). Most species of Petalidium occur in arid conditions where they receive no precipitation for at least 3 months during the driest quarter, yet the analysis suggests that the ancestor and early lineages of Petalidium (i.e., older nodes) occurred in more moist conditions. Thus, occupation of arid conditions appears to be a derived state. Right: Variation in net rate of diversification across Petalidium (Rabosky, 2014). This analysis suggests that one clade of Petalidium, that which encompasses the majority of species, is diversifying at a faster rate than the other clades in the phylogeny (with red, orange, and yellow branches). This clade also tends to occur in more arid conditions, and is referred to as the diverse arid clade in the main text.
In the BAMM analysis, the best model was one with one shift in diversification rate (posterior probability = 0.6), which is placed at the base of the diverse arid clade (Figure 4). This clade shows a higher net diversification rate than the rest of the phylogeny. When visualising model-averaged variation in net diversification across the phylogeny, we find that estimated net diversification rate ranged from 0.6 to 1.5 species per myrs.
With our QuaSSE analysis we found that diversification rate in Petalidium was best modelled as linearly related to PC1 (the “precipitation” component), with positive values (reflecting lower levels of precipitation, and presumably water availability) being related to higher net-diversification. Net diversification was negatively related to precipitation of the driest quarter, with highest net diversification in the most dry conditions. However, our simulations show that these relationships are not significantly different from what would be expected by chance. Indeed, the simulated ΔLL was greater than observed ΔLL 8% of the time for PC1 (p = 0.08) and 44% of the time for precipitation of driest quarter (p = 0.44). Overall, these results indicate that while there is a link between water availability and net diversification rate, with higher net diversification in arid regions, this link is not significant in a statistical sense.
Discussion
In this study, we found that the higher species richness of Petalidium in arid areas relative to moist areas may indeed be linked to higher rates of net diversification in arid areas, in contrast to the alternative hypothesis of a longer history of occupancy in arid areas. Our analyses suggest that the genus as a whole originated in more moist conditions ca 5 Ma and that two lineages subsequently shifted to occurring in more arid conditions ca 1.6 Ma (Figure 4). One lineage in particular, the diverse arid clade, showed elevated rates of net diversification, and this clade occupies the most arid areas within the overall range of the genus. Yet, the singular nature of the diverse arid clade prevents us from making strong causal links between occupation of arid areas and net diversification rate within the genus Petalidium. However, combining the results here for Petalidium with published results for other clades suggests there indeed may be a causal link (Fisher et al., 2015; Darbyshire et al., 2020).
An evolutionary pattern of higher diversification rates in arid areas during the Pleistocene has been found in other lineages of Acanthaceae in the Namib Desert and surrounding regions (Fisher et al., 2015; Darbyshire et al., 2020). Similarly, the majority of the species-rich lineages in the neighbouring Succulent Karoo arid biome are younger than 10 Ma (Klak et al., 2004; Verboom et al., 2009; García-Aloy et al., 2017). Thus, Petalidium represents an additional data point, notably from a woody plant clade, supporting a growing body of evidence for rapid Quaternary diversifications of plants in the Namib Desert and surrounding regions. It is key to note though that the Namib Desert also houses incredibly old evolutionary lineages that are found nowhere else on earth, such as Welwitschia, which diverged from Gnetum in the Cretaceous (Hou et al., 2015) and Tiganophyton, an early diverging member of the Brassicaceae which originated in the Eocene (Hendriks et al., 2022).
How then do we explain the co-occurrence of recently radiated lineages and ancient depauperons (Donoghue and Sanderson, 2015) in the same space? We hypothesize that the answer lies in exceptional evolutionary turnover. Recent studies (Koenen et al., 2015; Pennington et al., 2015) have suggested that high diversity in tropical rain forests can be explained by the persistence of numerous evolutionary lineages through episodic periods of extinction, which then provide ample ‘seed lineages’ from which radiations can arise. If episodes of extinction in arid biomes are more severe, then fewer lineages would persist throughout these episodes and fewer would radiate afterwards. Overall, plant richness would be lower in arid compared to moist biomes (as observed), but there nonetheless could still be multiple lineages that show recent diversification.
There is extensive evidence that the Namib desert experienced climatic fluctuations during the Quaternary with alternating arid and mesic phases, or pluvial cycles (Scott et al., 1991, 2022; Lancaster, 2002; Dupont, 2006; Gil-Romera et al., 2006; Brook et al., 2007; Grodek et al., 2013; Greenbaum et al., 2014; Lim et al., 2016). These oscillating mesic–arid phases could have provided opportunities for isolation, but also opportunities for previously isolated populations to exchange genes and produce novel genetic combinations, with both processes contributing to diversification, as suggested by Stebbins (1952). Furthermore, high levels of inter-annual variability in rainfall and extended droughts may also drive vegetation die back (Crouchet et al., 2019; Wright et al., 2023), leaving a “clean slate” available for colonisation during mesic periods. In fact, in our most recent fieldwork in the Namib Desert and surrounding areas, we observed extensive dieback of plant populations compared to our first observations in the early 2010s, presumably as a result of widespread drought during the last decade.
Our molecular dataset of thousands of SNPs was sufficient to resolve the deepest nodes in the phylogeny of Petalidium, but not relationships within the diverse arid clade, which shows low bootstrap support values (Figure 2, Supplementary Figure S1). Such low support was not expected given that RADseq has shown sufficient power to resolve fine-scale phylogenetic relationships in Barleria, a closely related genus of Acanthaceae (Comito et al., 2022). This poor resolution in the shallow part of our phylogeny could be explained by hybridisation or incomplete lineage sorting, both of which may cause conflict among genes for relationships. Although field observations (all authors, pers. obs.) document the existence of putative hybrids in Petalidium, i.e., individuals with morphology intermediate between two co-occurring species, a preliminary test for hybridisation with our SNP data did not suggest any evidence for hybridisation (Supplementary Figure S2). Given the young age (1.63 Ma) and the high species richness of the diverse arid clade, incomplete lineage sorting could be a plausible explanation for the lack of support, but new genetic data, from full genomes or numerous unlinked nuclear loci (c.f. Morais et al., 2019), will be necessary to fully address this issue in Petalidium.
Future phylogeny construction based on unlinked nuclear loci, rather than SNP data, may also allow for the implementation of better algorithms for temporally calibrating phylogenies (e.g., Mirarab et al., 2014). We relied on penalized likelihood to calibrate our ML phylogeny, a method that is known to bias node ages towards older time (Carruthers and Scotland, 2022). Nevertheless, our estimates for the ages of younger clades in the phylogeny, such as the diverse arid clade are similar to Tripp et al. (2017), suggesting that future improvements to the phylogeny will be unlikely to invalidate our finding that the majority of diversification in Petalidium, particularly in the diverse arid clade, occurred during the Quaternary.
Our findings show a clear correlation between adaptation to extreme aridity and increased net diversification in Petalidium, albeit not a statistically significant one. We have suggested that alternating mesic and arid phases in a present-day arid region have driven this pattern, but there are alternative explanations. The genus exhibits a remarkable diversity of floral forms, associated with different pollinators, including open cup-shaped flowers (a generalist pollination syndrome), large tubular red flowers (bird pollinated), and small maroon flowers (pollinated by long-tongued flies), and the diverse arid clade has the greatest floral diversity in Petalidium. Interactions with pollinators is a long-hypothesized driver of angiosperm diversity (Van der Niet et al., 2014; Tripp et al., 2021), and changes in floral form linked to switching of pollinators and subsequent reproductive isolation may have promoted speciation in Petalidium. Pollinators do seem scarce in this arid region at present, but they may have been less so during past mesic phases. Clearly, further investigation is needed to unravel the potentially interacting factors that promoted such a recent and fast diversification in one of the driest environments on Earth.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: NCBI; PRJNA930345.
Author contributions
OL, EM-T, and KGD conceived the study. EM-T and KGD collected the samples in the field that underlie the phylogeny. EM-T executed laboratory analyses. OL and EM-T conducted bioinformatic and phylogenetic analyses. WS contributed key insights into identification of specimens and phylogenetic results. OL and KGD conducted downstream macroevolutionary analyses and wrote the first draft of the article. All authors contributed to the article and approved the submitted version.
Funding
OL received funding from the Swiss National Science Foundation (P2LAP3_191377 and P500PB_203081). EM-T gratefully acknowledges support from the National Science Foundation Award # 0919594. EM-T further acknowledges extensive support from the University of Colorado Herbarium (COLO) staff, in particular Dina Clark and Ryan Allen, for assistance with processing herbarium materials resulting from her fieldwork. KGD warmly thanks the British Ecological Society, the Systematics Association, the Moray Endowment Fund of the University of Edinburgh and the Natural Environment Research Council (NERC) of the UK via the SECO Large Grant NE/T01279X/1. EM-T and KGD acknowledge the National Commission on Research, Science and Technology of Namibia and the Ministério da Agricultura e Florestas of Angola for permission to conduct field research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1193728/full#supplementary-material
References
Andrews, D. P.. (2017). FastQC: a quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc
Arakaki, M., Christin, P.-A., Nyffeler, R., Lendel, A., Eggli, U., Ogburn, R. M., et al. (2011). Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc. Natl. Acad. Sci. U.S.A. 108, 8379–8384. doi: 10.1073/pnas.1100628108
Aronesty, E. (2011). ea-utils: “command-line tools for processing biological sequence data”. Available at: https://github.com/ExpressionAnalysis/ea-utils
Axelrod, D. I. (1972). Edaphic aridity as a factor in angiosperm evolution. Am. Nat. 106, 311–320. doi: 10.1086/282773
Böhnert, T., Luebert, F., Merklinger, F. F., Harpke, D., Stoll, A., Schneider, J. V., et al. (2022). Plant migration under long-lasting hyperaridity–phylogenomics unravels recent biogeographic history in one of the oldest deserts on earth. New Phytol. 234, 1863–1875. doi: 10.1111/nph.18082
Böhnert, T., Luebert, F., Ritter, B., Merklinger, F. F., Stoll, A., Schneider, J. V., et al. (2019). Origin and diversification of Cristaria (Malvaceae) parallel Andean orogeny and onset of hyperaridity in the Atacama Desert. Glob. Planet. Chang. 181:102992. doi: 10.1016/j.gloplacha.2019.102992
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Brook, G. A., Marais, E., Srivastava, P., and Jordan, T. (2007). Timing of lake-level changes in Etosha Pan, Namibia, since the middle Holocene from OSL ages of relict shorelines in the Okondeka region. Quat. Int. 175, 29–40. doi: 10.1016/j.quaint.2007.05.020
Buira, A., Fernández-Mazuecos, M., Aedo, C., and Molina-Venegas, R. (2021). The contribution of the edaphic factor as a driver of recent plant diversification in a Mediterranean biodiversity hotspot. J. Ecol. 109, 987–999. doi: 10.1111/1365-2745.13527
Čalasan, A. Ž., Hammen, S., Sukhorukov, A. P., McDonald, J. T., Brignone, N. F., Böhnert, T., et al. (2022). From continental Asia into the world: global historical biogeography of the saltbush genus Atriplex (Chenopodieae, Chenopodioideae, Amaranthaceae). Perspect. Plant Ecol. Evol. Systemat. 54:125660. doi: 10.1016/j.ppees.2022.125660
Carruthers, T., and Scotland, R. W. (2022). exTREEmaTIME: a method for incorporating uncertainty into divergence time estimates. Biol. Open 11:bio059181. doi: 10.1242/bio.059181
Comito, R., Darbyshire, I., Kiel, C., McDade, L., and Fisher, A. E. (2022). A RADseq phylogeny of Barleria (Acanthaceae) resolves fine-scale relationships. Mol. Phylogenet. Evol. 169:107428. doi: 10.1016/j.ympev.2022.107428
Craven, P. (2009). Phytogeographic study of the Kaokoveld Centre of endemism. Doctoral dissertation, Stellenbosch: University of Stellenbosch.
Crouchet, S. E., Jensen, J., Schwartz, B. F., and Schwinning, S. (2019). Tree mortality after a hot drought: distinguishing density-dependent and-independent drivers and why it matters. Front. For. Glob. Change 2:21. doi: 10.3389/ffgc.2019.00021
Currie, D. J. (1991). Energy and large-scale patterns of animal-and plant-species richness. Am. Nat. 137, 27–49. doi: 10.1086/285144
Darbyshire, I., Kiel, C. A., Astroth, C. M., Dexter, K. G., Chase, F. M., and Tripp, E. A. (2020). Phylogenomic study of Monechma reveals two divergent plant lineages of ecological importance in the african savanna and succulent biomes. Diversity 12:237. doi: 10.3390/d12060237
Daru, B. H., Van Der Bank, M., Maurin, O., Yessoufou, K., Schaefer, H., Slingsby, J. A., et al. (2016). A novel phylogenetic regionalization of phytogeographical zones of southern Africa reveals their hidden evolutionary affinities. J. Biogeogr. 43, 155–166. doi: 10.1111/jbi.12619
Donoghue, M. J., and Sanderson, M. J. (2015). Confluence, synnovation, and depauperons in plant diversification. New Phytol. 207, 260–274. doi: 10.1111/nph.13367
Doyle, J. J., and Doyle, J. L. (1987). A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Dupont, L. M. (2006). Late Pliocene vegetation and climate in Namibia (southern Africa) derived from palynology of ODP site 1082. Geochem. Geophys. Geosyst. 7, 1–15. doi: 10.1029/2005GC001208
Eaton, D. A. R., and Overcast, I. (2020). Ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594. doi: 10.1093/bioinformatics/btz966
Esquivel-Muelbert, A., Baker, T. R., Dexter, K. G., Lewis, S. L., Ter Steege, H., Lopez-Gonzalez, G., et al. (2017). Seasonal drought limits tree species across the Neotropics. Ecography 40, 618–629. doi: 10.1111/ecog.01904
Felsenstein, J. (1985). Phylogenies and the comparative method. Am. Nat. 125, 1–15. doi: 10.1086/284325
Fick, S. E., and Hijmans, R. J. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. doi: 10.1002/joc.5086
Fisher, A. E., McDade, L. A., Kiel, C. A., Khoshravesh, R., Johnson, M. A., Stata, M., et al. (2015). Evolutionary history of Blepharis (Acanthaceae) and the origin of C4 photosynthesis in section Acanthodium. Int. J. Plant Sci. 176, 770–790. doi: 10.1086/683011
FitzJohn, R. G. (2010). Quantitative traits and diversification. Syst. Biol. 59, 619–633. doi: 10.1093/sysbio/syq053
FitzJohn, R. G. (2012). Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092. doi: 10.1111/j.2041-210X.2012.00234.x
García-Aloy, S., Sanmartín, I., Kadereit, G., Vitales, D., Millanes, A. M., Roquet, C., et al. (2017). Opposite trends in the genus Monsonia (Geraniaceae): specialization in the African deserts and range expansions throughout eastern Africa. Sci. Rep. 7, 9872–9813. doi: 10.1038/s41598-017-09834-6
Gil-Romera, G., Scott, L., Marais, E., and Brook, G. A. (2006). Middle-to late-Holocene moisture changes in the desert of Northwest Namibia derived from fossil hyrax dung pollen. The Holocene 16, 1073–1084. doi: 10.1177/0959683606069397
Greenbaum, N., Schwartz, U., Benito, G., Porat, N., Cloete, G. C., and Enzel, Y. (2014). Paleohydrology of extraordinary floods along the Swakop River at the margin of the Namib Desert and their paleoclimate implications. Quat. Sci. Rev. 103, 153–169. doi: 10.1016/j.quascirev.2014.08.021
Grodek, T., Benito, G., Botero, B. A., Jacoby, Y., Porat, N., Haviv, I., et al. (2013). The last millennium largest floods in the hyperarid Kuiseb River basin, Namib Desert. J. Quat. Sci. 28, 258–270. doi: 10.1002/jqs.2618
Guerrero, P. C., Rosas, M., Arroyo, M. T. K., and Wiens, J. J. (2013). Evolutionary lag times and recent origin of the biota of an ancient desert (Atacama–Sechura). Proc. Natl. Acad. Sci. U.S.A. 110, 11469–11474. doi: 10.1073/pnas.1308721110
Harmon, L. J., Weir, J. T., Brock, C. D., Glor, R. E., and Challenger, W. (2007). GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131. doi: 10.1093/bioinformatics/btm538
Hendriks, K. P., Kiefer, C., Al-Shehbaz, I. A., Bailey, C. D., Hooft van Huysduynen, A., Nikolov, L. A., et al. (2022). Global phylogeny of the Brassicaceae provides important insights into gene discordance. bioRxiv, 2022-09.
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hou, C., Humphreys, A. M., Thureborn, O., and Rydin, C. (2015). New insights into the evolutionary history of Gnetum (Gnetales). Taxon 64, 239–253. doi: 10.12705/642.12
Hua, X., Cardillo, M., and Bromham, L. (2022). Adapting to extremes: reconstructing evolution in response to changing climate over time and space in the diverse Australian plant genus Acacia. J. Biogeogr. 49, 727–738. doi: 10.1111/jbi.14339
Kadereit, J. W., and Abbott, R. J. (2021). Plant speciation in the quaternary. Plant Ecol. Divers. 14, 105–142. doi: 10.1080/17550874.2021.2012849
Klak, C., Reeves, G., and Hedderson, T. (2004). Unmatched tempo of evolution in southern African semi-desert ice plants. Nature 427, 63–65. doi: 10.1038/nature02243
Koenen, E., Clarkson, J. J., Pennington, T., and Chatrou, L. W. (2015). Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol. 207, 327–339. doi: 10.1111/nph.13490
Lancaster, N. (2002). How dry was dry?—late Pleistocene palaeoclimates in the Namib Desert. Quat. Sci. Rev. 21, 769–782. doi: 10.1016/S0277-3791(01)00126-3
Lavor, P., Calvente, A., Versieux, L. M., and Sanmartin, I. (2019). Bayesian spatio-temporal reconstruction reveals rapid diversification and Pleistocene range expansion in the widespread columnar cactus Pilosocereus. J. Biogeogr. 46, 238–250. doi: 10.1111/jbi.13481
Leaché, A. D., Banbury, B. L., Felsenstein, J., de Oca, A. N.-M., and Stamatakis, A. (2015). Short tree, long tree, right tree, wrong tree: new acquisition Bias corrections for inferring SNP phylogenies. Syst. Biol. 64, 1032–1047. doi: 10.1093/sysbio/syv053
Lim, S., Chase, B. M., Chevalier, M., and Reimer, P. J. (2016). 50,000years of vegetation and climate change in the southern Namib Desert, Pella, South Africa. Palaeogeogr. Palaeoclimatol. Palaeoecol. 451, 197–209. doi: 10.1016/j.palaeo.2016.03.001
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Merklinger, F. F., Böhnert, T., Arakaki, M., Weigend, M., Quandt, D., and Luebert, F. (2021). Quaternary diversification of a columnar cactus in the driest place on earth. Am. J. Bot. 108, 184–199. doi: 10.1002/ajb2.1608
Mirarab, S., Reaz, R., Bayzid, M. S., Zimmermann, T., Swenson, M. S., and Warnow, T. (2014). ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548. doi: 10.1093/bioinformatics/btu462
Morais, E. B., Schönenberger, J., Conti, E., Antonelli, A., and Szövényi, P. (2019). Orthologous nuclear markers and new transcriptomes that broadly cover the phylogenetic diversity of Acanthaceae. Appl. Plant Sci. 7:e11290. doi: 10.1002/aps3.11290
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
O'Brien, E. M., Field, R., and Whittaker, R. J. (2000). Climatic gradients in woody plant (tree and shrub) diversity: water-energy dynamics, residual variation, and topography. Oikos 89, 588–600. doi: 10.1034/j.1600-0706.2000.890319.x
Parchman, T. L., Gompert, Z., Mudge, J., Schilkey, F. D., Benkman, C. W., and Buerkle, C. A. (2012). Genome-wide association genetics of adaptive traits in lodgepole pine. Mol. Ecol. 21, 2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x
Pennell, M. W., Eastman, J. M., Slater, G. J., Brown, J. W., Uyeda, J. C., FitzJohn, R. G., et al. (2014). Geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics 30, 2216–2218. doi: 10.1093/bioinformatics/btu181
Pennington, R. T., Hughes, M., and Moonlight, P. W. (2015). The origins of tropical rainforest hyperdiversity. Trends Plant Sci. 20, 693–695. doi: 10.1016/j.tplants.2015.10.005
Pennington, R. T., Richardson, J. E., and Lavin, M. (2006). Insights into the historical construction of species-rich biomes from dated plant phylogenies, neutral ecological theory and phylogenetic community structure. New Phytol. 172, 605–616. doi: 10.1111/j.1469-8137.2006.01902.x
Plummer, M., Best, N., Cowles, K., and Vines, K. (2008). Coda: output analysis and diagnostics for MCMC. R package version 0.13–3. Available at: http://CRAN.R-project.org/package=coda
R Core Team (2013). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Rabosky, D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9:e89543. doi: 10.1371/journal.pone.0089543
Rabosky, D. L., and Goldberg, E. E. (2015). Model inadequacy and mistaken inferences of trait-dependent speciation. Syst. Biol. 64, 340–355. doi: 10.1093/sysbio/syu131
Rabosky, D. L., Grundler, M., Anderson, C., Title, P., Shi, J. J., Brown, J. W., et al. (2014). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707. doi: 10.1111/2041-210X.12199
Revell, L. J. (2012). Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Scott, L., Cooremans, B., de Wet, J. S., and Vogel, J. C. (1991). Holocene environmental changes in Namibia inferred from pollen analysis of swamp and lake deposits. The Holocene 1, 8–13. doi: 10.1177/095968369100100103
Scott, L., Gil-Romera, G., Marais, E., and Brook, G. A. (2022). Holocene environmental change along the central Namib Desert escarpment derived from hyrax and owl dung. Rev. Palaeobot. Palynol. 305:104746. doi: 10.1016/j.revpalbo.2022.104746
Simon, M. F., Grether, R., de Queiroz, L. P., Skema, C., Pennington, R. T., and Hughes, C. E. (2009). Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proc. Natl. Acad. Sci. U.S.A. 106, 20359–20364. doi: 10.1073/pnas.0903410106
Singhal, S., Roddy, A. B., DiVittorio, C., Sanchez-Amaya, A., Henriquez, C. L., Brodersen, C. R., et al. (2021). Diversification, disparification and hybridization in the desert shrubs Encelia. New Phytol. 230, 1228–1241. doi: 10.1111/nph.17212
Smith, S. A., and O’Meara, B. C. (2012). treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. doi: 10.1093/bioinformatics/bts492
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stebbins, G. L. (1952). Aridity as a stimulus to plant evolution. Am. Nat. 86, 33–44. doi: 10.1086/281699
Swanepoel, W. (2020). Petalidium kaokoense (Acanthaceae), a new species from Namibia. Phytotaxa 468, 236–242. doi: 10.11646/phytotaxa.468.3.1
Swanepoel, W., Chase, M. W., Christenhusz, M. J., Maurin, O., Forest, F., and van Wyk, A. E. (2020). From the frying pan: an unusual dwarf shrub from Namibia turns out to be a new brassicalean family. Phytotaxa 439, 171–185. doi: 10.11646/phytotaxa.439.3.1
Swanepoel, W., and Manzitto-Tripp, E. A. (2022). Petalidium sesfonteinense (Acanthaceae), a new species from the Kaokoveld, Namibia. Phytotaxa 549, 127–135. doi: 10.11646/phytotaxa.549.2.1
Tripp, E. A., Dexter, K. G., and Stone, H. B. (2021). Reproductive character displacement and potential underlying drivers in a species-rich and florally diverse lineage of tropical angiosperms (Ruellia; Acanthaceae). Ecol. Evol. 11, 4719–4730. doi: 10.1002/ece3.7371
Tripp, E. A., Tsai, Y.-H. E., Zhuang, Y., and Dexter, K. G. (2017). RADseq dataset with 90% missing data fully resolves recent radiation of Petalidium (Acanthaceae) in the ultra-arid deserts of Namibia. Ecol. Evol. 7, 7920–7936. doi: 10.1002/ece3.3274
Van der Niet, T., Peakall, R., and Johnson, S. D. (2014). Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Ann. Bot. 113, 199–212. doi: 10.1093/aob/mct290
Verboom, G. A., Archibald, J. K., Bakker, F. T., Bellstedt, D. U., Conrad, F., Dreyer, L. L., et al. (2009). Origin and diversification of the greater cape flora: ancient species repository, hot-bed of recent radiation, or both? Mol. Phylogenet. Evol. 51, 44–53. doi: 10.1016/j.ympev.2008.01.037
Ward, J. D., Seely, M. K., and Lancaster, N. (1983). On the antiquity of the Namib. S. Afr. J. Sci. 79, 175–183.
Wright, S. (1940). Breeding structure of populations in relation to speciation. Am. Nat. 74, 232–248. doi: 10.1086/280891
Wright, B. R., Nipper, M., Nipper, N., Merson, S. D., and Guest, T. (2023). Mortality rates of desert vegetation during high-intensity drought at Ulu ru-Kata Tju ta National Park, Central Australia. Austral Ecol. 1–20. doi: 10.1111/aec.13290
Keywords: Namib Desert, arid biomes, Acanthaceae, plant diversification, RAD sequencing
Citation: Loiseau O, Manzitto-Tripp EA, Swanepoel W and Dexter KG (2023) Net diversification rates of the woody plant genus Petalidium (Acanthaceae) are highest in the ancient and arid Namib Desert. Front. Ecol. Evol. 11:1193728. doi: 10.3389/fevo.2023.1193728
Edited by:
Danilo T. Amaral, Federal University of ABC, BrazilReviewed by:
Milena Telhe, Federal University of São Carlos, BrazilMarcos Queiroz, Institute of Botany (ASCR), Czechia
Copyright © 2023 Loiseau, Manzitto-Tripp, Swanepoel and Dexter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle G. Dexter, kyle.dexter@ed.ac.uk
 Oriane Loiseau
Oriane Loiseau Erin A. Manzitto-Tripp
Erin A. Manzitto-Tripp Wessel Swanepoel3
Wessel Swanepoel3  Kyle G. Dexter
Kyle G. Dexter