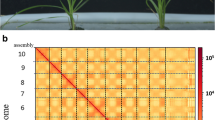
Abstract
Guar (Cyamopsis tetragonoloba (L.) Taub.) is becoming a popular industrial crop in response to industry demand for the guar gum extracted from seeds’ endosperm. Breeding efforts of new guar varieties would greatly benefit from genomic resources developed for marker assisted selection (MAS) purposes. We have undertaken the first steps to establish a whole-genome assembly of the guar ‘Vaviloskij 130’ accession, bred at VIR. Using a combination of second (Illumina short reads) and third generation (Oxford Nanopore long reads) sequencing methods, a dataset of approx. 5X of genome coverage was obtained. We tested assemblers for short reads, namely SOAPdenovo, AbySS and SGA, based on different algorithms. For short reads (Illumina MiSeq and HiSeq data), the better result in terms of total number of scaffolds and total assembly length were obtained with SGA (String Graph Assembler). For Oxford Nanopore dataset, we used the combination of minimap + miniASM assembly, then corrected the assembly with raw Illumina and Nanopore data. The current preliminary de novo assembly of the guar genome covers 1.2 Gb, corresponding to 50% of the genome. The data confirm the phylogeny position of C. tetragoloba as being highly related to the genus Vigna, Abrus, Glycine and Lupinus genomes. This preliminary reference genome paves the way to further detailed diversity and genetic analyses into that important agro-industrial crop.




Similar content being viewed by others
REFERENCES
Aswathnarayana, N.K., Tiwari, P.B., Choudhary, M., et al., Genetic diversity study of cluster bean (Cyamopsis tetragonoloba (L.) Taub.) landraces using RAPD and ISSR markers, Int. J. Adv. Biotechnol. Res., 2013, vol. 4, no. 4, pp. 460—471.
Mudgil, D., Barak, S., and Khatkar, B.S., Guar gum: processing, properties and food applications—a review, J. Food Sci. Technol., 2014, vol. 51, no. 3, pp. 409—418. https://doi.org/10.1007/s13197-011-0522-x
Lubbers, E.L., Characterization and inheritance of photoperiodism on guar, Cyamopsis tetragonoloba (L.) Taub., PhD Thesis, University of Arizona. 1987.
Whistler, R.L. and Hymowitz, T., Guar: Agronomy, Production, Industrial Use, and Nutrition, Purdue University Press, 1979.
Douglas, C.A., Evaluation of Guar Cultivars in Central and Southern Queensland: A Report for the Rural Industries Research and Development Corporation, Barton, Australia: Rural Industries Research and Development Corporation, 2005.
Boghara, M.C., Dhaduk, H.L., Kumar, et al., Genetic divergence, path analysis and molecular diversity analysis in cluster bean (Cyamopsis tetragonoloba L. Taub.), Ind. Crops Prod., 2016, vol. 89, pp. 468—477. https://doi.org/10.15389/agrobiology.2017.6.1116eng
Gresta, F., Avola, G., Cannavò, S., et al., Morphological, biological, productive and qualitative characterization of 68 guar (Cyamopsis tetragonoloba (L.) Taub.) genotypes, Ind. Crops Prod., 2018, vol. 114, pp. 98—107. https://doi.org/10.1016/j.indcrop.2018.01.070
Dzyubenko, N.I., Dzyubenko, E.A., Potokina, E.K., et al., Clusterbeans Cyamopsis tetragonoloba (L.) Taub.—properties, use, plant genetic resources and expected introduction in Russia, S.-kh. Biol., vol. 52, no. 6, pp. 1116—1128. https://doi.org/10.15389/agrobiology.2017.6.1116rus
Kumar, S., Modi, A.R., Parekh, M.J., et al., Role of conventional and biotechnological approaches for genetic improvement of cluster bean, Ind. Crops Prod., 2017, vol. 97, pp. 639—648. https://doi.org/10.1016/j.indcrop.2017.01.008
Naoumkina, M., Torres-Jerez, I., Allen, S., et al., Analysis of cDNA libraries from developing seeds of guar (Cyamopsis tetragonoloba (L.) Taub.), BMC Plant Biol., 2007, vol. 7, no. 1, p. 62. https://doi.org/10.1186/1471-2229-7-62
Rawal, H.C., Kumar, S., Mithra, S.V.A., et al., High quality unigenes and microsatellite markers from tissue specific transcriptome and development of a database in clusterbean (Cyamopsis tetragonoloba L. Taub.), Genes, 2017, vol. 8, no. 11, p. 313. https://doi.org/10.3390/genes8110313
Tanwar, U.K., Pruthi, V., Randhawa, G.S., RNA-Seq of guar (Cyamopsis tetragonoloba L. Taub.) leaves: de novo transcriptome assembly, functional annotation and development of genomic resources, Front. Plant Sci., 2017, vol. 8, p. 91. https://doi.org/10.3389/fpls.2017.0009
Thakur, O. and Randhawa, G.S., Identification and characterization of SSR, SNP and InDel molecular markers from RNA-Seq data of guar (Cyamopsis tetragonoloba L. Taub.) roots, BMC Genomics, 2018, vol. 19, no. 1, p. 951. https://doi.org/10.1186/s12864-018-5205-9
Sahu, S., Rao, A.R., Pandey, J., et al., Genome-wide identification and characterization of lncRNAs and miRNAs in cluster bean (Cyamopsis tetragonoloba), Gene, 2018, vol. 667, pp. 112—12. https://doi.org/10.1016/j.gene.2018.05.027
Tyagi, A., Nigam, D., Solanke, A.U., et al., Genome-wide discovery of tissue-specific miRNAs in clusterbean (Cyamopsis tetragonoloba) indicates their association with galactomannan biosynthesis, Plant Biotechnol. J., 2018, vol. 16, no. 6, pp. 1241—1257. https://doi.org/10.1111/pbi.12866
Kaila, T., Chaduvla, P.K., Rawal, et al., Chloroplast genome sequence of clusterbean (Cyamopsis tetragonoloba L.): genome structure and comparative analysis, Genes, 2017, vol. 8, no. 9, p. 212. https://doi.org/10.3390/genes8090212
Patil, C.G., Nuclear DNA amount variation in Cyamopsis D.C. (Fabaceae), Cytologia, 2004, vol. 69, no. 1, pp. 59—62. https://doi.org/10.1508/cytologia.69.59
Tyagi, A., Sandhya, S., Sharma, et al., The genome size of clusterbean (Cyamopsis tetragonoloba) is significantly smaller compared to its wild relatives as estimated by flow cytometry, Gene, 2019. https://doi.org/10.1111/pbi.12866
Chang, Y., Liu, H., Liu, M., et al., The draft genomes of five agriculturally important African orphan crops, GigaScience, 2018, vol. 8, no. 3, р. 152. https://doi.org/10.1093/gigascience/giy152
Bennett, M.D., Smith J.B., and Riley Ralph, Nuclear DNA amounts in angiosperms, R. Soc. Online J., 1976, vol. 274, pp. 227—274. https://doi.org/10.1098/rspb.1982.0069
Metzker, M.L., Sequencing technologies—the next generation, Nat. Rev. Genet., 2010, vol. 11, no. 1, p. 31. https://doi.org/10.1038/nrg2626
van Dijk, E.L., Auger, H., Jaszczyszyn, Y., et al., Ten years of next-generation sequencing technology, Trends Genet., 2014, vol. 30, no. 9, pp. 418—426. https://doi.org/10.1016/j.tig.2014.07.001
Ip, C.L.C., Loose, M., Tyson, J.R., et al., MinION Analysis and Reference Consortium: phase 1 data release and analysis, F1000Research, 2015, vol. 4. https://doi.org/10.12688/f1000research.7201.1
de Lannoy, C., de Ridder, D., and Risse, J., The long reads ahead: de novo genome assembly using the MinION, F1000Research, 2017, vol. 6. https://doi.org/10.12688/f1000research.12012.2
Simpson, J.T. and Pop, M., The theory and practice of genome sequence assembly, Annu. Rev. Genomics Hum. Genet., 2015, vol. 16, pp. 153—172. https://doi.org/10.1146/annurev-genom-090314-050032
Myers, E.W., Sutton, G.G., Delcher, A.L., et al., A whole-genome assembly of Drosophila,Science, 2000, vol. 287, no. 5461, pp. 2196—2204.
Compeau, P.E.C., Pevzner, P.A., and Tesler, G., Why are de Bruijn graphs useful for genome assembly?, Nat. Biotechnol., 2011, vol. 29, no. 11, p. 987. https://doi.org/10.1038/nbt.2023
Myers, E.W., The fragment assembly string graph, Bioinformatics, 2005, vol. 21, suppl. 2, pp. ii79—ii85. https://doi.org/10.1093/bioinformatics/bti1114
Pevzner, P.A., Tang, H., and Waterman, M.S., An Eulerian path approach to DNA fragment assembly, Proc. Natl. Acad. Sci., U.S.A., 2001, vol. 98, no. 17, pp. 9748—9753. https://doi.org/10.1073/pnas.171285098
Simpson, J.T. and Durbin, R., Efficient construction of an assembly string graph using the FM-index, Bioinformatics, 2010, vol. 26, no. 12, pp. i367—i373. https://doi.org/10.1093/bioinformatics/btq217
Doyle, J., A rapid total DNA preparation procedure for fresh plant tissue, Focus, 1990, vol. 12, pp. 13—15. https://doi.org/10.1007/978-3-642-83962-7_18
FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
BBTools. https://jgi.doe.gov/data-and-tools/bbtools/.
Luo, R., Liu, B., Xie, Y., et al., SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler, GigaScience, 2012, vol. 1, p. 18. https://doi.org/10.1186/2047-217X-1-18
Jackman, S.D., Vandervalk, B.P., Mohamadi, H., et al., ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter, Genome Res., 2017, vol. 27, no. 5, pp. 768—777. https://doi.org/10.1101/gr.214346.116
Simpson, J.T. and Durbin, R., Efficient de novo assembly of large genomes using compressed data structures, Genome Res., 2012, vol. 22, no. 3, pp. 549—556. https://doi.org/10.1101/gr.126953.111
De Coster, W., D’Hert, S., Schultz, D.T., et al., Bioinformatics, 2018, vol. 34, no. 15, pp. 2666—2669. https://doi.org/10.1093/bioinformatics/bty149
Li, H., Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences, Bioinformatics, 2016, vol. 32, no. 14, pp. 2103—2110. https://doi.org/10.1093/bioinformatics/btw152
Walker, B.J., Abeel, T., Shea, T., et al., Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement, PLoS One, 2014, vol. 9, no. 11. e112963. https://doi.org/10.1371/journal.pone.0112963
Antipov, D., Korobeynikov, A., McLean, J.S., et al., hybridSPAdes: an algorithm for hybrid assembly of short and long reads, Bioinformatics, 2015, vol. 32, no. 7, pp. 1009—1015. https://doi.org/10.1093/bioinformatics/btv68
Gurevich, A., Saveliev, V., Vyahhi Tehler, G., et al., QUAST: quality assessment tool for genome assemblies, Bioinformatics, 2013, vol. 29, no. 8, pp. 1072—1075. https://doi.org/10.1093/bioinformatics/btt086
Zhang, Z., Schwartz, S., Wagner, L., et al., Greedy algorithm for aligning DNA sequences, J. Comput. Biol., 2000, vol. 7, nos. 1—2, pp. 203—214. https://doi.org/10.1089/10665270050081478
Cock, P.J.A., Antao, T., Chang, J.T., et al., Biopython: freely available Python tools for computational molecular biology and bioinformatics, Bioinformatics, 2009, vol. 25, no. 11, pp. 1422—1423. https://doi.org/10.1093/bioinformatics/btp163
R Core Team, R: A Language and Environment for Statistical Computing, Vienna: R Foundation for Statistical Computing, 2019.
Zimin, A.V., Marçais, G., Puiu, D., et al., The MaSuRCA genome assembler, Bioinformatics, 2013, vol. 29, no. 21, pp. 2669—2677. https://doi.org/10.1093/bioinformatics/btt476
Peters, W.S., Haffer, D., Hanakam, C.B., et al., Legume phylogeny and the evolution of a unique contractile apparatus that regulates phloem transport, Am. J. Bot., 2010, vol. 97, no. 5, pp. 797—808. https://doi.org/10.3732/ajb.0900328
Pratap, A. and Kumar, J., Biology and Breeding of Food Legumes, CABI, 2011. https://doi.org/10.1017/S0014479712000312
Wang, J., Sun, P., Li, Y., Cheng, R., Duan, X., et al., Hierarchically aligning 10 legume genomes establishes a family-level genomics platform, Plant Physiol., 2017, vol. 174, no. 1, pp. 284—300. https://doi.org/10.1104/pp.16.01981
Belser, C., Istace, B., Denis, E., et al., Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps, Nat. Plants, 2018, vol. 4, no. 11, p. 879. https://doi.org/10.1038/s41477-018-0289-4
Schmidt, M.H.-W., Vogel, A., Denton, A.K., et al., De novo assembly of a new Solanum pennellii accession using nanopore sequencing, plant cell, Plant Cell, 2017, vol. 29, no. 10, pp. 2336—2348. https://doi.org/10.1105/tpc.17.00521
Chaisson, M.J.P., Wilson, R.K., and Eichler, E.E., Genetic variation and the de novo assembly of human genomes, Nat. Rev. Genet., 2015, vol. 16, no. 11, p. 627. https://doi.org/10.1038/nrg3933
Ellinghaus, D., Kurtz, S., and Willhoeft, U., LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons, BMC Bioinf., 2008, vol. 9, no. 1, p. 18. https://doi.org/10.1186/1471-2105-9-18
RepeatMasker Open-4.0. http://repeatmasker.org.
Sohn, J. and Nam, J.-W., The present and future of de novo whole-genome assembly, Briefings Bioinf., 2016, vol. 19, no. 1, pp. 23—40. https://doi.org/10.1093/bib/bbw096
ACKNOWLEDGMENTS
The reported study was funded by RFBR according to the research project no. 17-29-08027. We are grateful to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing computing resources. Sequencing was performed at the Biobank of Research Park of St. Petersburg State University (no. 1259502495). E. Potokina was supported by a “Visiting Scholar” fellowship from Toulouse INP for a short scientific stay at Toulouse INP.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Grigoreva, E., Ulianich, P., Ben, C. et al. First Insights into the Guar (Cyamopsis tetragonoloba (L.) Taub.) Genome of the ‘Vavilovskij 130’ Accession, Using Second and Third-Generation Sequencing Technologies. Russ J Genet 55, 1406–1416 (2019). https://doi.org/10.1134/S102279541911005X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S102279541911005X