Abstract
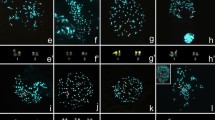
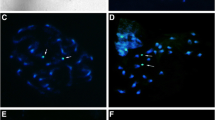
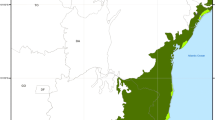
Nassella is one of the largest genera in the tribe Stipeae with approximately 116 species, distributed from Canada to Uruguay and Argentina. Although there are strong morphological characters that distinguish this genus from the rest of the tribe, molecular phylogenetic analyses have shown that it is a polyphyletic genus closely related to Jarava and Amelichloa. Species boundaries for many Nassella taxa are somewhat diffuse and controversial, mainly due to morphological similarities with just a few morphological characters that differentiate one from the other. Previous chromosome counts in Nassella have shown the existence of a high chromosome number variability ranging from 28 to 60 suggesting that polyploidy might have played an important role in the evolution of the genus. The aim of this study was to assess the relationship between chromosome number variation and genome size focusing in 15 species of Nassella distributed in Uruguay and to establish the relationship among N. megapotamia (Spreng. ex Trin.) Barkworth, N. pauciciliata (Roseng. & Izag.) Barkworth, N. poeppigiana (Trin. & Rupr.) Barkworth and N. quinqueciliata (Roseng. & Izag.) Barkworth & Torres using cytogenetic, morphological, and molecular phylogenetic data. Chromosome numbers ranged from 2n = 26 to 2n = 46 confirming the high chromosome number variability reported for the tribe. This is the first record of nuclear DNA content for the genus and 2C-values range from 1.48 to 2.31 pg representing a 1.5-fold difference. There is no direct correspondence between increase in chromosome number and DNA content. This lack of correspondence could be due to DNA loss after polyploidization, or related to chromosomal rearrangements. Four Nassella species exhibited high cytological and morphological homogeneity. Our data based on chromosome number, molecular phylogenetic analysis, and the morphological approach including both optical and scanning electron microscopy strongly support that these four Nassella species (N. megapotamia, N. pauciciliata, N. poeppigiana, N. quinqueciliata) form a complex of species whose boundaries should be revised.






Similar content being viewed by others
References
Barber J, Hames KA, Cialdella AM, Giussani LM, Morrone O (2009) Phylogenetic relationships of Piptochaetium Presl (Poaceae: Stipeae) and related genera reconstructed from nuclear and chloroplast sequence datasets. Taxon 58:375–380
Barkworth ME (1983) Ptilagrostis in North America and its relationship to other stipeae (Gramineae). Syst Bot 8:395–419
Barkworth ME (1990) Nassella (Gramineae, Stipeae): revised interpretation and nomenclatural changes. Taxon 39:597–614
Barkworth ME, Torres MA (2001) Distribution and diagnostic characters of Nassella (Poaceae: Stipeae). Taxon 50:439–468
Barkworth ME, Arriaga MO, Smith JF, Jacobs SWL, Valdé-Reyna J, Bushman BS (2008) Molecules and morphology in South American Stipeae (Poaceae). Syst Bot 33:719–731
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot Lond 107:467–590
Bowden WM, Senn HA (1962) Chromosome numbers in 28 grass genera from South America. Can J Bot 40:1115–1124
Bureš P, Zedek F (2014) Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution 68:2412–2420
Cialdella AM (2012) Tribu Stipeae. In: Zuloaga FO, Rúgolo de Agrasar ZE, Anton AM (eds) Flora vascular de la República Argentina, vol 3(2). Gráficamente Ediciones, Córdoba, pp 372–495
Cialdella AM, Giussani LM, Aagesen L, Zuloaga FO, Morrone O (2007) A Phylogeny of Piptochaetium (Poaceae: Pooideae: Stipeae) and related genera based on a combined analysis including Trnl-f, Rpl16, and morphology. Syst Bot 32:545–559
Cialdella AM, Salariato DL, Aagesen L, Giussani LM, Zuloaga FO, Morrone O (2010) Phylogeny of New World Stipeae (Poaceae): an evaluation of the monophyly of Aciachne and Amelichloa. Cladistics 26:563–578
Cialdella AM, Muñoz-Schick M, Morrone O (2013) Sinopsis de las especies austro-americanas del género Nassella. Darwiniana Nueva Ser 1:76–161
Cialdella AM, Sede SM, Romaschenko K, Peterson PM, Soreng RJ, Zuloaga FO, Morrone O (2014) Phylogeny of Nassella (Stipeae, Pooideae, Poaceae) based on analyses of chloroplast and nuclear ribosomal DNA and morphology. Syst Bot 39:814–828
Clayton WD, Renvoize SA (1986) Genera graminum. Grasses of the world. Kew Bull. Additional Series XIII, Royal Botanic Gardens, Kew
Doležel J, Göhde W (1995) Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19:103–106
Doležel J, Sgorbati S, Lucretti S (1992) Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plant 85:625–631
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Elias MK (1942) Tertiary prairie grasses and other herbs from the high plains. Geol Soc Am Spec Pap 41:1–176
González AC (2011) Stipa L. (s.l.) en el Uruguay. Revisión del Género y Análisis de su distribución geográfica. Dissertation, Universidad de la República, Montevideo
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23:1029–1041
Guerra M (2012) Cytotaxonomy: the end of childhood. Plant Biosyst 146:703–710
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Jacobs SWL, Everett J, Barkworth ME (1995) Clarification of morphological terms used in the Stipeae (Gramineae), and a reassessment of Nassella in Australia. Taxon 44:33–41
Johnson BL (1972) Polyploidy as a factor in the evolution of grasses. In: Younger B, McKell CV (eds) The biology and utilization of grasses. Academic Press, New York, pp 18–35
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc 82:651–663
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
Lipnerová I, Bureš P, Horová L, Smarda P (2013) Evolution of genome size in Carex (Cyperaceae) in relation to chromosome number and genomic base composition. Ann Bot Lond 111:79–94
Myers WM (1947) Cytology and genetics of forage grasses. Bot Rev 13:319–421
Romaschenko K, Peterson PM, Soreng RJ, Garcia-Jacas N, Futorna O, Susanna A (2008) Molecular phylogenetic analysis of the American Stipeae (Poaceae) resolves Jarava sensu lato polyphyletic: evidence for a new genus, Pappostipa. J Bot Res Inst Tex 2:165–192
Romaschenko K, Peterson PM, Soreng RJ, Garcia-Jacas N, Susanna A (2010) Phylogenetics of Stipeae (Poaceae: Pooideae) based on plastid and nuclear DNA sequences. In: Seberg O, Petersen G, Barfod A, Davis JI (eds) Diversity, phylogeny, and evolution in the monocotyledons, 10th edn. Aarhus University Press, Denmark, pp 511–537
Romaschenko K, Peterson PM, Soreng RJ, Garcia-Jacas N, Futorna O, Susanna A (2012) Systematics and evolution of the needle grasses (Poaceae: Pooideae: Stipeae) based on analysis of multiple chloroplast loci, ITS, and lemma micromorphology. Taxon 61:18–44
Romaschenko K, Garcia-Jacas N, Peterson PM, Soreng RJ, Vilatersana R, Susanna A (2014) Miocene–Pliocene speciation, introgression, and migration of Patis and Ptilagrostis (Poaceae: Stipeae). Mol Phylogenet Evol 70:244–259
Rosengurtt B, Arrillaga MB, Izaguirre P (1970) Gramíneas uruguayas. Universidad de la República, Montevideo
Saura R (1943) Cariología de gramíneas: géneros Paspalum, Stipa, Poa, Andropogon y Phalaris. Inst. Genet. Fac. Agron. Vet. Buenos Aires 2:65–74
Speranza P, Vaio M, Mazzella C (2003) Karyotypes of two cytotypes of Paspalum quadrifarium Lam. (Poaceae): an alternative technique for small chromosomes in plants. Genet Mol Biol 26:499–503
Vaio M, Speranza P, Valls JFM, Guerra M, Mazzella C (2005) Localization of the 45S and 5S rDNA and cpDNA sequence analysis in species of the Quadrifaria group of Paspalum (Poaceae, Paniceae). Ann Bot Lond 96:191–200
Vaio M, Gardner A, Emshwiller E, Guerra M (2013) Molecular phylogeny and chromosome evolution among the creeping herbaceous Oxalis species of sections Corniculatae and Ripariae (Oxalidaceae). Mol Phylogenet Evol 68:199–211
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Winterfeld G, Schneider J, Becher H, Dickie J, Röser M (2015) Karyosystematics of the Australasian stipoid grass Austrostipa and related genera: chromosome sizes, ploidy, chromosome base numbers and phylogeny. Aust Syst Bot 28:145–159
Zuloaga F, Morrone O, Belgrano M (2008) Catálogo de las plantas vasculares del Cono Sur. Argentina, Sur de Brasil, Chile, Paraguay y Uruguay. Vol 1. Pteridophyta, Monocotyledonae, Gymnospermae. Instituto Darwinion, Buenos Aires
Acknowledgements
This work was supported by grants from Comisión Sectorial de Investigación Científica and PEDECIBA (Universidad de la República, Uruguay). We are indebted to Professor Primavera Izaguirre for her advice in the dissertation thesis of A. González, Pablo Speranza for his invaluable comments that improved the manuscript, Graciela Ververis for botanical illustrations, and B. López-Carro and L. Mondos for technical assistance. We wish to thank J. Doležel for providing the Zea mays seeds.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
González, A.C., Vaio, M., Porro, V. et al. Chromosome numbers, DNA content, morphological data, and nrITS sequence analyses in some species of Nassella (Trin.) E. Desv. and related genera (Stipeae, Poaceae). Braz. J. Bot 40, 341–352 (2017). https://doi.org/10.1007/s40415-016-0337-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-016-0337-0